6OK2
 
 | |
6OLK
 
 | |
6OJX
 
 | |
6OKV
 
 | |
6OJY
 
 | |
6OLL
 
 | |
6OLJ
 
 | |
5TSH
 
 | | PilB from Geobacter metallireducens bound to AMP-PNP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | McCallum, M, Tammam, S, Khan, A, Burrows, L, Howell, P.L. | | 登録日 | 2016-10-28 | | 公開日 | 2017-05-17 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The molecular mechanism of the type IVa pilus motors.
Nat Commun, 8, 2017
|
|
5TSG
 
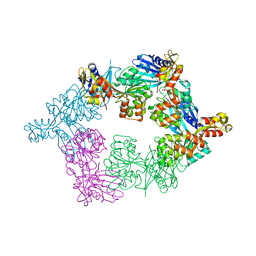 | | PilB from Geobacter metallireducens bound to ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, FORMIC ACID, MAGNESIUM ION, ... | | 著者 | McCallum, M, Tammam, S, Khan, A, Burrows, L, Howell, P.L. | | 登録日 | 2016-10-28 | | 公開日 | 2017-05-17 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.4011 Å) | | 主引用文献 | The molecular mechanism of the type IVa pilus motors.
Nat Commun, 8, 2017
|
|
7TLY
 
 | |
7TLZ
 
 | |
7TN0
 
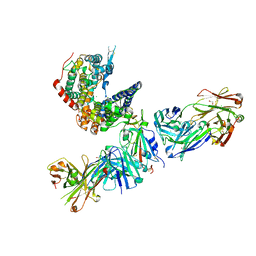 | | SARS-CoV-2 Omicron RBD in complex with human ACE2 and S304 Fab and S309 Fab | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | McCallum, M, Czudnochowski, N, Nix, J.C, Croll, T.I, SSGCID, Dillen, J.R, Snell, G, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2022-01-20 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement.
Science, 375, 2022
|
|
7TM0
 
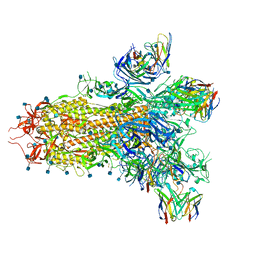 | |
6VE4
 
 | | Pentadecameric PilQ from Pseudomonas aeruginosa | | 分子名称: | Fimbrial assembly protein PilQ | | 著者 | McCallum, M, Tammam, S, Rubinstein, J.L, Burrows, L.L, Howell, P.L. | | 登録日 | 2019-12-28 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (6.9 Å) | | 主引用文献 | CryoEM map of Pseudomonas aeruginosa PilQ enables structural characterization of TsaP.
Structure, 29, 2021
|
|
6VE2
 
 | | Tetradecameric PilQ bound by TsaP heptamer from Pseudomonas aeruginosa | | 分子名称: | Fimbrial assembly protein PilQ, LysM domain-containing protein | | 著者 | McCallum, M, Tammam, S, Rubinstein, J.L, Burrows, L.L, Howell, P.L. | | 登録日 | 2019-12-28 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | CryoEM map of Pseudomonas aeruginosa PilQ enables structural characterization of TsaP.
Structure, 29, 2021
|
|
6VE3
 
 | | Tetradecameric PilQ from Pseudomonas aeruginosa | | 分子名称: | Fimbrial assembly protein PilQ | | 著者 | McCallum, M, Tammam, S, Rubinstein, J.L, Burrows, L.L, Howell, P.L. | | 登録日 | 2019-12-28 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | CryoEM map of Pseudomonas aeruginosa PilQ enables structural characterization of TsaP.
Structure, 29, 2021
|
|
8VYE
 
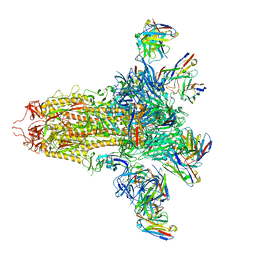 | | SARS-CoV-2 S (C.37 Lambda variant) plus S309, S2L20, and S2X303 Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S2L20 Heavy Chain, S2L20 Light Chain, ... | | 著者 | McCallum, M, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2024-02-08 | | 公開日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Quantifying how single dose Ad26.COV2.S vaccine efficacy depends on Spike sequence features.
Nat Commun, 15, 2024
|
|
8VYG
 
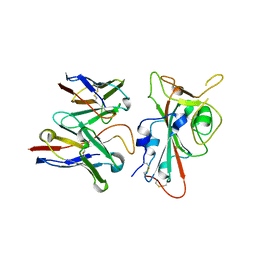 | | SARS-CoV-2 S RBD (C.37 Lambda variant) plus S309 Fab, local refinement | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S309 Heavy Chain, S309 Light Chain, ... | | 著者 | McCallum, M, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2024-02-08 | | 公開日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Quantifying how single dose Ad26.COV2.S vaccine efficacy depends on Spike sequence features.
Nat Commun, 15, 2024
|
|
8VYF
 
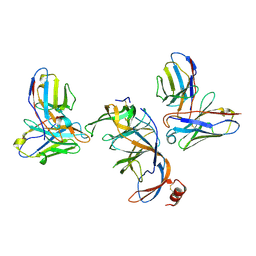 | | SARS-CoV-2 S NTD (C.37 Lambda variant) plus S2L20 and S2X303 Fabs, local refinement | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2L20 Heavy Chain, ... | | 著者 | McCallum, M, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2024-02-08 | | 公開日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Quantifying how single dose Ad26.COV2.S vaccine efficacy depends on Spike sequence features.
Nat Commun, 15, 2024
|
|
8DF5
 
 | | SARS-CoV-2 Beta RBD in complex with human ACE2 and S304 Fab and S309 Fab | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | McCallum, M, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | 登録日 | 2022-06-21 | | 公開日 | 2022-08-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Shifting mutational constraints in the SARS-CoV-2 receptor-binding domain during viral evolution.
Science, 377, 2022
|
|
8TVI
 
 | |
8VGT
 
 | |
8VWP
 
 | |
7SL5
 
 | |
7SKZ
 
 | |
