9F9A
 
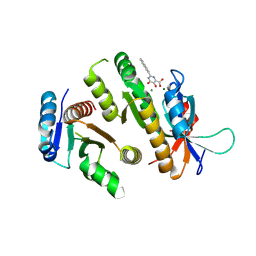 | | Crystal structure of MUS81-EME1 bound by compound 12. | | 分子名称: | 2-naphthalen-2-yl-5-oxidanyl-6-oxidanylidene-1H-pyrimidine-4-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | 著者 | Collie, G.W. | | 登録日 | 2024-05-07 | | 公開日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.911 Å) | | 主引用文献 | Fragment-Based Discovery of Novel MUS81 Inhibitors
Acs Med.Chem.Lett., 2024
|
|
8HKG
 
 | |
6NW9
 
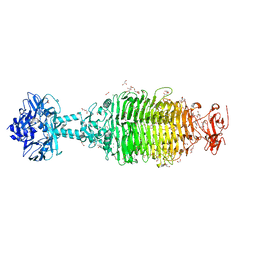 | | CRYSTAL STRUCTURE OF A TAILSPIKE PROTEIN 3 (TSP3, ORF212) FROM ESCHERICHIA COLI O157:H7 BACTERIOPHAGE CBA120 | | 分子名称: | 1,2-ETHANEDIOL, CARBONATE ION, CHLORIDE ION, ... | | 著者 | Greenfield, J.Y, Herzberg, O. | | 登録日 | 2019-02-06 | | 公開日 | 2019-06-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure and tailspike glycosidase machinery of ORF212 from E. coli O157:H7 phage CBA120 (TSP3).
Sci Rep, 9, 2019
|
|
6A50
 
 | | structure of benzoylformate decarboxylases in complex with cofactor TPP | | 分子名称: | MAGNESIUM ION, THIAMINE DIPHOSPHATE, benzoylformate decarboxylases | | 著者 | Guo, Y, Wang, S, Nie, Y, Li, S. | | 登録日 | 2018-06-21 | | 公開日 | 2019-02-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A Synthetic Pathway for Acetyl-Coenzyme A Biosynthesis
Nat Commun, 2019
|
|
7WVR
 
 | |
5AY7
 
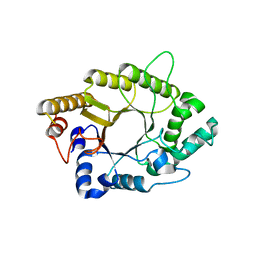 | | A psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase | | 分子名称: | xylanase | | 著者 | Zheng, Y, Li, Y, Liu, W, Guo, R.T. | | 登録日 | 2015-08-10 | | 公開日 | 2016-02-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural insight into potential cold adaptation mechanism through a psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase.
J.Struct.Biol., 193, 2016
|
|
8K6C
 
 | |
8K6D
 
 | |
8K68
 
 | |
8K6A
 
 | |
8K67
 
 | |
8K6B
 
 | |
5OFL
 
 | | Crystal structure of CbXyn10C variant E140Q/E248Q complexed with cellohexaose | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Glycoside hydrolase family 48, SULFATE ION, ... | | 著者 | Hakulinen, N, Penttinen, L, Rouvinen, J. | | 登録日 | 2017-07-11 | | 公開日 | 2017-10-04 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.871 Å) | | 主引用文献 | Insights into the roles of non-catalytic residues in the active site of a GH10 xylanase with activity on cellulose.
J. Biol. Chem., 292, 2017
|
|
5OFK
 
 | | Crystal structure of CbXyn10C variant E140Q/E248Q complexed with xyloheptaose | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Glycoside hydrolase family 48, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), ... | | 著者 | Hakulinen, N, Penttinen, L, Rouvinen, J, Tu, T. | | 登録日 | 2017-07-11 | | 公開日 | 2017-10-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Insights into the roles of non-catalytic residues in the active site of a GH10 xylanase with activity on cellulose.
J. Biol. Chem., 292, 2017
|
|
5OFJ
 
 | | Crystal structure of N-terminal domain of bifunctional CbXyn10C | | 分子名称: | 1,2-ETHANEDIOL, CITRATE ANION, Glycoside hydrolase family 48 | | 著者 | Hakulinen, N, Penttinen, L, Rouvinen, J. | | 登録日 | 2017-07-11 | | 公開日 | 2017-10-04 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Insights into the roles of non-catalytic residues in the active site of a GH10 xylanase with activity on cellulose.
J. Biol. Chem., 292, 2017
|
|
3NDQ
 
 | | Structure of Human TFIIS Domain II | | 分子名称: | SODIUM ION, Transcription elongation factor A protein 1 | | 著者 | Hu, X.P, Averell, G, Ye, X.Y, Hou, J, Luo, H. | | 登録日 | 2010-06-07 | | 公開日 | 2011-05-18 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.933 Å) | | 主引用文献 | Structure of Human TFIIS Domain II
To be Published
|
|
6KVE
 
 | |
5XZO
 
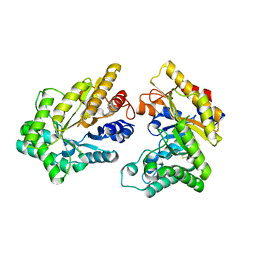 | | Crystal structure of GH10 xylanase XYL10C from Bispora. sp MEY-1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-xylanase | | 著者 | You, S, Chen, C, Tu, T, Guo, R.T, Luo, H, Yao, B. | | 登録日 | 2017-07-13 | | 公開日 | 2018-01-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of GH10 xylanase XYL10C from Bispora. sp MEY-1
To Be Published
|
|
3WUB
 
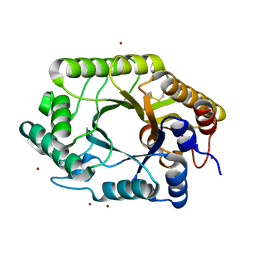 | | The wild type crystal structure of b-1,4-Xylanase (XynAS9) from Streptomyces sp. 9 | | 分子名称: | Endo-1,4-beta-xylanase A, ZINC ION | | 著者 | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y.Y, Guo, R.T. | | 登録日 | 2014-04-23 | | 公開日 | 2014-10-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
3WUG
 
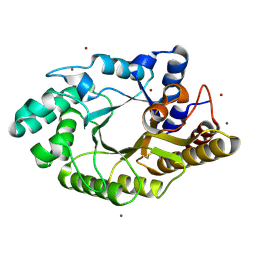 | | The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) with xylobiose from Streptomyces sp. 9 | | 分子名称: | Endo-1,4-beta-xylanase A, ZINC ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | 著者 | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y.Y, Guo, R.T. | | 登録日 | 2014-04-23 | | 公開日 | 2014-10-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
3WUE
 
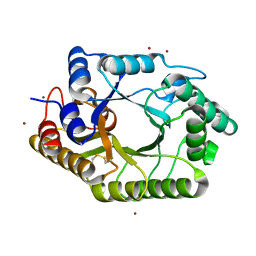 | | The wild type crystal structure of b-1,4-Xylanase (XynAS9) with xylobiose from Streptomyces sp. 9 | | 分子名称: | Endo-1,4-beta-xylanase A, ZINC ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | 著者 | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y, Guo, R.T. | | 登録日 | 2014-04-23 | | 公開日 | 2014-10-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
3WUF
 
 | | The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) from Streptomyces sp. 9 | | 分子名称: | Endo-1,4-beta-xylanase A, ZINC ION | | 著者 | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y.Y, Guo, R.T. | | 登録日 | 2014-04-23 | | 公開日 | 2014-10-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
3V36
 
 | | Aldose reductase complexed with glceraldehyde | | 分子名称: | Aldose reductase, D-Glyceraldehyde, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Zheng, X, Zhang, L, Chen, Y, Luo, H, Hu, X. | | 登録日 | 2011-12-13 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Partial inhibition of aldose reductase by nitazoxanide and its molecular basis.
Chemmedchem, 7, 2012
|
|
3V35
 
 | | Aldose reductase complexed with a nitro compound | | 分子名称: | 2-[(5-nitro-1,3-thiazol-2-yl)carbamoyl]phenyl acetate, Aldose reductase, DIMETHYLFORMAMIDE, ... | | 著者 | Zheng, X, Zhang, L, Chen, Y, Luo, H, Hu, X. | | 登録日 | 2011-12-13 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Partial inhibition of aldose reductase by nitazoxanide and its molecular basis.
Chemmedchem, 7, 2012
|
|
