8BUD
 
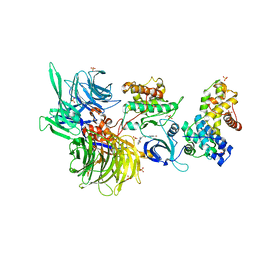 | | Structure of DDB1 bound to Z7-engaged CDK12-cyclin K | | 分子名称: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUE
 
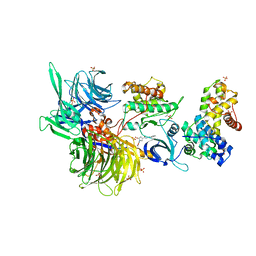 | | Structure of DDB1 bound to Z11-engaged CDK12-cyclin K | | 分子名称: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUG
 
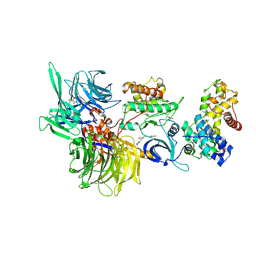 | | Structure of DDB1 bound to HQ461-engaged CDK12-cyclin K | | 分子名称: | 2-[2-[(6-methylpyridin-2-yl)amino]-1,3-thiazol-4-yl]-~{N}-(5-methyl-1,3-thiazol-2-yl)ethanamide, CITRIC ACID, Cyclin-K, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.53 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
6TD3
 
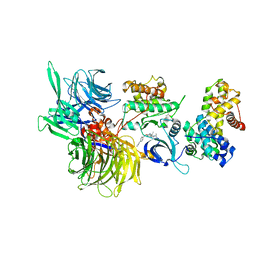 | | Structure of DDB1 bound to CR8-engaged CDK12-cyclinK | | 分子名称: | (2R)-2-({9-(1-methylethyl)-6-[(4-pyridin-2-ylbenzyl)amino]-9H-purin-2-yl}amino)butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Bunker, R.D, Petzold, G, Kozicka, Z, Thoma, N.H. | | 登録日 | 2019-11-07 | | 公開日 | 2020-06-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.46 Å) | | 主引用文献 | The CDK inhibitor CR8 acts as a molecular glue degrader that depletes cyclin K.
Nature, 585, 2020
|
|
6YOV
 
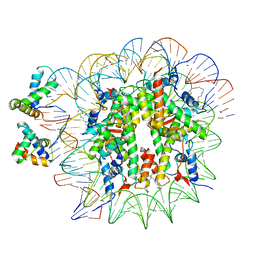 | | OCT4-SOX2-bound nucleosome - SHL+6 | | 分子名称: | DNA (142-MER), Green fluorescent protein,POU domain, class 5, ... | | 著者 | Michael, A.K, Kempf, G, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2020-04-15 | | 公開日 | 2020-05-06 | | 最終更新日 | 2020-07-08 | | 実験手法 | ELECTRON MICROSCOPY (3.42 Å) | | 主引用文献 | Mechanisms of OCT4-SOX2 motif readout on nucleosomes.
Science, 368, 2020
|
|
6Y5D
 
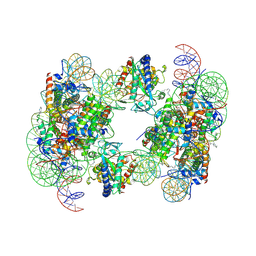 | | Structure of human cGAS (K394E) bound to the nucleosome | | 分子名称: | Cyclic GMP-AMP synthase, DNA (153-MER), Histone H2A type 2-A, ... | | 著者 | Pathare, G.R, Cavadini, S, Kempf, G, Thoma, N.H. | | 登録日 | 2020-02-25 | | 公開日 | 2020-09-23 | | 最終更新日 | 2020-12-09 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural mechanism of cGAS inhibition by the nucleosome.
Nature, 587, 2020
|
|
6Y5E
 
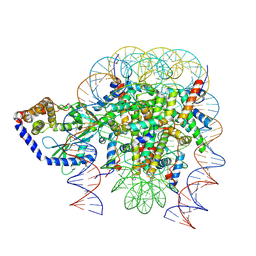 | | Structure of human cGAS (K394E) bound to the nucleosome (focused refinement of cGAS-NCP subcomplex) | | 分子名称: | Cyclic GMP-AMP synthase, DNA (153-MER), Histone H2A type 2-C, ... | | 著者 | Pathare, G.R, Cavadini, S, Kempf, G, Thoma, N.H. | | 登録日 | 2020-02-25 | | 公開日 | 2020-09-23 | | 最終更新日 | 2020-12-09 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Structural mechanism of cGAS inhibition by the nucleosome.
Nature, 587, 2020
|
|
6T90
 
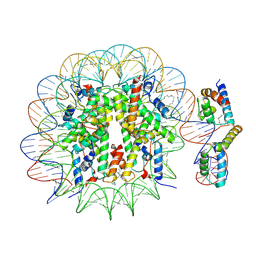 | | OCT4-SOX2-bound nucleosome - SHL-6 | | 分子名称: | DNA (146-MER), Green fluorescent protein,POU domain, class 5, ... | | 著者 | Michael, A.K, Kempf, G, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-10-25 | | 公開日 | 2020-05-06 | | 最終更新日 | 2020-07-08 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Mechanisms of OCT4-SOX2 motif readout on nucleosomes.
Science, 368, 2020
|
|
6T93
 
 | | Nucleosome with OCT4-SOX2 motif at SHL-6 | | 分子名称: | DNA (153-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Michael, A.K, Kempf, G, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-10-25 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | Mechanisms of OCT4-SOX2 motif readout on nucleosomes.
Science, 368, 2020
|
|
