6UAH
 
 | | Crystal Structure of the Metallo-beta-Lactamase L1 from Stenotrophomonas maltophilia in the Complex with Hydrolyzed Meropenem | | 分子名称: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ... | | 著者 | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-09-10 | | 公開日 | 2019-09-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Crystal Structure of the Metallo-beta-Lactamase L1 from Stenotrophomonas maltophilia in the Complex with Hydrolyzed Meropenem
To Be Published
|
|
8EBC
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Listeria monocytogenes in the complex with IMP | | 分子名称: | FORMIC ACID, GLYCEROL, INOSINIC ACID, ... | | 著者 | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Osipiuk, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2022-08-31 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Listeria monocytogenes in the complex with IMP
To Be Published
|
|
7N3C
 
 | | Crystal Structure of Human Fab S24-202 in the complex with the N-terminal Domain of Nucleocapsid protein from SARS CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, IODIDE ION, Nucleoprotein, ... | | 著者 | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-05-31 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
7N3D
 
 | | Crystal Structure of Human Fab S24-1564 in the complex with the N-terminal Domain of Nucleocapsid protein from SARS CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Nucleoprotein, ... | | 著者 | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-05-31 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
3C5O
 
 | | Crystal structure of the conserved protein of unknown function RPA1785 from Rhodopseudomonas palustris | | 分子名称: | GLYCEROL, UPF0311 protein RPA1785 | | 著者 | Kim, Y, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-02-01 | | 公開日 | 2008-02-19 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Crystal Structure of the Conserved Protein of Unknown Function RPA1785 from Rhodopseudomonas palustris.
To be Published
|
|
3EFB
 
 | | Crystal Structure of Probable sor Operon Regulator from Shigella flexneri | | 分子名称: | ACETIC ACID, Probable sor-operon regulator | | 著者 | Kim, Y, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2008-09-08 | | 公開日 | 2008-09-23 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Crystal Structure of Probable sor Operon Regulator from Shigella flexneri
To be Published
|
|
3EC7
 
 | | Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 | | 分子名称: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETIC ACID, ... | | 著者 | Kim, Y, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-08-29 | | 公開日 | 2008-09-23 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2
To be Published
|
|
6XPG
 
 | | Crystal Structure of Sialate O-acetylesterase from Bacteroides vulgatus by Serial Crystallography | | 分子名称: | Lysophospholipase L1 | | 著者 | Kim, Y, Sherrell, D.A, Owen, R, Axford, D, Ebrahim, A, Johnson, J, Welk, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2020-07-08 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of Sialate O-acetylesterase from Bacteroides vulgatus by Serial Crystallography
To Be Published
|
|
6XPM
 
 | | Crystal Structure of Sialate O-acetylesterase from Bacteroides vulgatus with microfluidics crystals at room temperature | | 分子名称: | Lysophospholipase L1, SODIUM ION | | 著者 | Kim, Y, Johnson, J, Welk, L, Endres, M, Levens, A, Sherrell, D.A, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2020-07-08 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structure of Sialate O-acetylesterase from Bacteroides vulgatus with microfluidics crystals at room temperature
To Be Published
|
|
8CRV
 
 | | Crystal Structure of the Carbamate Kinase from Pseudomonas aeruginosa | | 分子名称: | 1,2-ETHANEDIOL, Carbamate kinase, FORMIC ACID, ... | | 著者 | Kim, Y, Skarina, T, Mesa, N, Stogios, P, Savchenko, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-05-11 | | 公開日 | 2022-08-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structure of the Carbamate Kinase from Pseudomonas aeruginosa
To Be Published
|
|
7TOC
 
 | | Crystal Structure of the Mitochondrial Ketol-acid Reductoisomerase IlvC from Candida auris | | 分子名称: | ACETIC ACID, Ketol-acid reductoisomerase, mitochondrial, ... | | 著者 | Kim, Y, Evdokimova, E, Di, R, Stogios, P, Savchenko, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-01-24 | | 公開日 | 2022-02-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Crystal Structure of the Mitochondrial Ketol-acid Reductoisomerase IlvC from Candida auris
To Be Published
|
|
5TF0
 
 | | Crystal Structure of Glycosil Hydrolase Family 3 N-Terminal Domain Protein from Bacteroides intestinalis | | 分子名称: | 1,2-ETHANEDIOL, Glycosyl hydrolase family 3 N-terminal domain protein, MAGNESIUM ION | | 著者 | Kim, Y, Hatzos-Skintges, C, Endres, M, Babnigg, G, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2016-09-23 | | 公開日 | 2016-10-05 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.2021 Å) | | 主引用文献 | Crystal Structure of Glycosil Hydrolase Family 3 N-Terminal Domain Protein from Bacteroides intestinalis
To Be Published, 2016
|
|
5TK4
 
 | | Crystal Structure of Uncharacterized Cupredoxin-like Domain Protein from Bacillus anthracis | | 分子名称: | Cytochrome B | | 著者 | Kim, Y, Maltseva, N, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-10-06 | | 公開日 | 2016-11-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Crystal Structure of Uncharacterized Cupredoxin-like Domain Protein from
Bacillus anthracis
To Be Published
|
|
5TF3
 
 | | Crystal Structure of Protein of Unknown Function YPO2564 from Yersinia pestis | | 分子名称: | 1,2-ETHANEDIOL, Putative membrane protein | | 著者 | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Anderson, W.F, Crosson, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-09-23 | | 公開日 | 2016-10-19 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Crystal Structure of Protein of Unknown Function YPO2564 from Yersinia pestis
To Be Published
|
|
5TK2
 
 | | Crystal Structure of Uncharacterized Cupredoxin-like domain protein from Bacillus anthracis | | 分子名称: | 1,2-ETHANEDIOL, CADMIUM ION, Cytochrome B, ... | | 著者 | Kim, Y, Maltseva, N, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-10-06 | | 公開日 | 2016-11-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal Structure of Uncharacterized Cupredoxin-like domain protein from
Bacillus anthracis
To Be Published
|
|
1XPP
 
 | | Crystal Structure of TA1416,DNA-directed RNA polymerase subunit L, from Thermoplasma acidophilum | | 分子名称: | ACETIC ACID, DNA-directed RNA polymerase subunit L, FORMIC ACID, ... | | 著者 | Kim, Y, Joachimiak, A, Evdokimova, E, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2004-10-09 | | 公開日 | 2004-11-23 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of TA1416, DNA-directed RNA polymerase subunit L, from Thermoplasma acidophilum
To be Published
|
|
7CA3
 
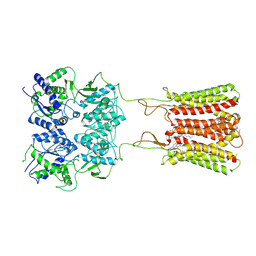 | | Cryo-EM structure of human GABA(B) receptor bound to the positive allosteric modulator rac-BHFF | | 分子名称: | (3S)-5,7-ditert-butyl-3-oxidanyl-3-(trifluoromethyl)-1-benzofuran-2-one, CHOLESTEROL, Gamma-aminobutyric acid type B receptor subunit 1, ... | | 著者 | Kim, Y, Jeong, E, Jeong, J, Kim, Y, Cho, Y. | | 登録日 | 2020-06-08 | | 公開日 | 2020-11-11 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structural Basis for Activation of the Heterodimeric GABA B Receptor.
J.Mol.Biol., 432, 2020
|
|
7CA5
 
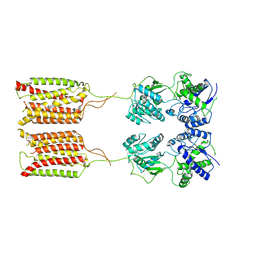 | | Cryo-EM structure of human GABA(B) receptor in apo state | | 分子名称: | Gamma-aminobutyric acid type B receptor subunit 1, Gamma-aminobutyric acid type B receptor subunit 2 | | 著者 | Kim, Y, Jeong, E, Jeong, J, Kim, Y, Cho, Y. | | 登録日 | 2020-06-08 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (7.6 Å) | | 主引用文献 | Structural Basis for Activation of the Heterodimeric GABA B Receptor.
J.Mol.Biol., 432, 2020
|
|
7CUM
 
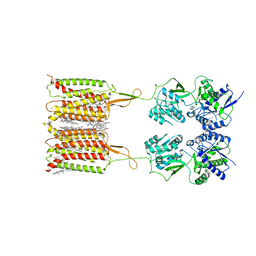 | | Cryo-EM structure of human GABA(B) receptor bound to the antagonist CGP54626 | | 分子名称: | (R)-(cyclohexylmethyl)[(2S)-3-{[(1S)-1-(3,4-dichlorophenyl)ethyl]amino}-2-hydroxypropyl]phosphinic acid, CHOLESTEROL, Gamma-aminobutyric acid type B receptor subunit 1, ... | | 著者 | Kim, Y, Jeong, E, Jeong, J, Kim, Y, Cho, Y. | | 登録日 | 2020-08-23 | | 公開日 | 2020-11-11 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Structural Basis for Activation of the Heterodimeric GABA B Receptor.
J.Mol.Biol., 432, 2020
|
|
2O38
 
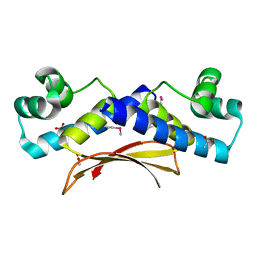 | | Putative XRE Family Transcriptional Regulator | | 分子名称: | ACETIC ACID, Hypothetical protein | | 著者 | Kim, Y, Joachimiak, A, Evdokimova, E, Kagan, O, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2006-11-30 | | 公開日 | 2007-01-02 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | The Crystal Structure of Putative XRE Family Transcriptional Regulator
To be Published
|
|
2O35
 
 | | Protein of Unknown Function (DUF1244) from Sinorhizobium meliloti | | 分子名称: | Hypothetical protein DUF1244, MAGNESIUM ION | | 著者 | Kim, Y, Joachimiak, A, Evdokimova, E, Kudritska, M, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2006-11-30 | | 公開日 | 2007-01-02 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | The Crystal Structure of Protein of Unknown Function (DUF1244) from Sinorhizobium meliloti
To be Published
|
|
2P12
 
 | | Crystal structure of protein of unknown function DUF402 from Rhodococcus sp. RHA1 | | 分子名称: | ACETIC ACID, GLYCEROL, Hypothetical protein DUF402 | | 著者 | Kim, Y, Evdokimova, E, Kudritska, M, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2007-03-01 | | 公開日 | 2007-04-03 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | The crystal structure of the protein of uncharacterized function, DUF402 from Rhodococcus sp. RHA1
To be Published
|
|
2G7G
 
 | | The Crystal Structure of the Putative Transcriptional Regulator Rha04620 from Rhodococcus sp. RHA1 | | 分子名称: | ACETIC ACID, Rha04620, Putative Transcriptional Regulator | | 著者 | Kim, Y, Joachimiak, A, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A.M, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2006-02-28 | | 公開日 | 2006-03-28 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | The Crystal Structure of the Putative Transcriptional Regulator Rha04620 from Rhodococcus sp. RHA1
To be Published
|
|
8VZE
 
 | | Crystal Structure of 2-Hydroxyacyl-CoA lyase/synthase TbHACS from Thermoflexaceae bacterium in the Complex with THDP, 2-Hydroxyisobutyryl-CoA and ADP | | 分子名称: | 1,2-ETHANEDIOL, 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-2-(hydroxymethyl)-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Kim, Y, Maltseva, M, Endres, M, Lee, S, Yoshikuni, Y, Gonzalez, R, Michalska, K, Joachimiak, A. | | 登録日 | 2024-02-11 | | 公開日 | 2024-10-02 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Revealing reaction intermediates in one-carbon elongation by thiamine diphosphate/CoA-dependent enzyme family.
Commun Chem, 7, 2024
|
|
8VZH
 
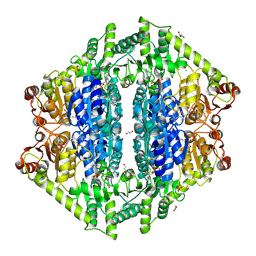 | | Crystal Structure of 2-Hydroxyacyl-CoA lyase/synthase TbHACS from Thermoflexaceae bacterium in the Complex with THDP and ADP | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | 著者 | Kim, Y, Maltseva, M, Endres, M, Lee, S, Yoshikuni, Y, Gonzalez, R, Michalska, K, Joachimiak, A. | | 登録日 | 2024-02-11 | | 公開日 | 2024-10-02 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Revealing reaction intermediates in one-carbon elongation by thiamine diphosphate/CoA-dependent enzyme family.
Commun Chem, 7, 2024
|
|
