6M37
 
 | |
6M36
 
 | |
5WUQ
 
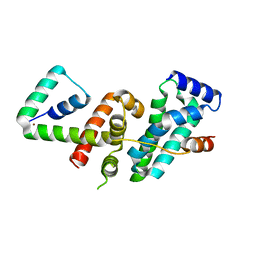 | | Crystal structure of SigW in complex with its anti-sigma RsiW, a zinc binding form | | 分子名称: | Anti-sigma-W factor RsiW, ECF RNA polymerase sigma factor SigW, ZINC ION | | 著者 | Devkota, S.R, Kwon, E, Ha, S.C, Kim, D.Y. | | 登録日 | 2016-12-20 | | 公開日 | 2017-03-29 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural insights into the regulation of Bacillus subtilis SigW activity by anti-sigma RsiW
PLoS ONE, 12, 2017
|
|
5WUR
 
 | | Crystal structure of SigW in complex with its anti-sigma RsiW, an oxdized form | | 分子名称: | Anti-sigma-W factor RsiW, ECF RNA polymerase sigma factor SigW | | 著者 | Devkota, S.R, Kwon, E, Ha, S.C, Kim, D.Y. | | 登録日 | 2016-12-20 | | 公開日 | 2017-03-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural insights into the regulation of Bacillus subtilis SigW activity by anti-sigma RsiW
PLoS ONE, 12, 2017
|
|
5X55
 
 | |
6JHE
 
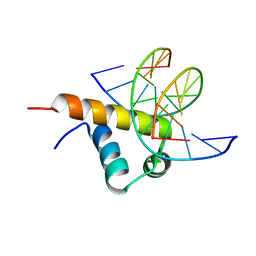 | | Crystal Structure of Bacillus subtilis SigW domain 4 in complexed with -35 element DNA | | 分子名称: | DNA (5'-D(*AP*AP*AP*GP*GP*TP*TP*TP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*AP*AP*AP*CP*CP*TP*TP*T)-3'), ECF RNA polymerase sigma factor SigW | | 著者 | Kwon, E, Devkota, S.R, Pathak, D, Dahal, P, Kim, D.Y. | | 登録日 | 2019-02-18 | | 公開日 | 2020-01-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.101 Å) | | 主引用文献 | Structural analysis of the recognition of the -35 promoter element by SigW from Bacillus subtilis.
Plos One, 14, 2019
|
|
6LYE
 
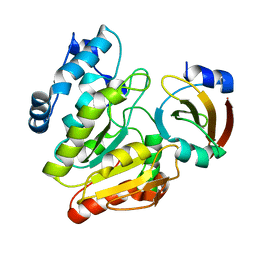 | |
6LYD
 
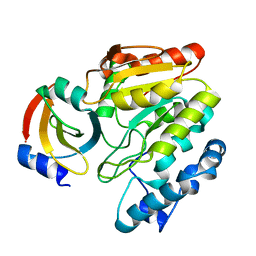 | |
7CX5
 
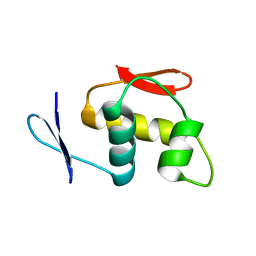 | |
7CJ2
 
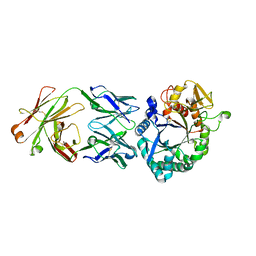 | | Crystal structure of the Fab antibody complexed with human YKL-40 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 3-like 1 (Cartilage glycoprotein-39), isoform CRA_a, ... | | 著者 | Choi, S, Na, J.H, Lee, S.J, Woo, J.R, Kim, D.Y, Hong, J.T, Lee, W.K. | | 登録日 | 2020-07-09 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of the Fab antibody complexed with human YKL-40
To Be Published
|
|
6C3R
 
 | |
1EFE
 
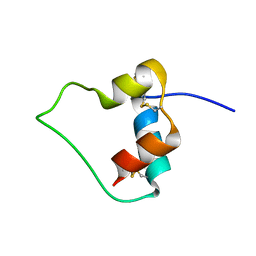 | | AN ACTIVE MINI-PROINSULIN, M2PI | | 分子名称: | MINI-PROINSULIN | | 著者 | Cho, Y, Chang, S.G, Choi, K.D, Shin, H, Ahn, B, Kim, K.S. | | 登録日 | 2000-02-08 | | 公開日 | 2000-03-17 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of an Active Mini-Proinsulin, M2PI: Inter-chain Flexibility is Crucial for Insulin Activity
J.Biochem.Mol.Biol., 33, 2000
|
|
8Z9B
 
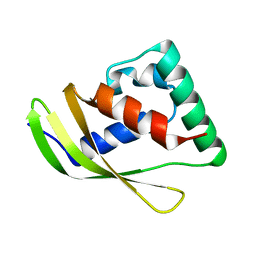 | | Low molecular weight antigen MTB12 | | 分子名称: | Low molecular weight antigen MTB12 | | 著者 | Park, H.H, Han, J.H. | | 登録日 | 2024-04-23 | | 公開日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Novel structure of secreted small molecular weight antigen Mtb12 from Mycobacterium tuberculosis.
Biochem.Biophys.Res.Commun., 717, 2024
|
|
5LS0
 
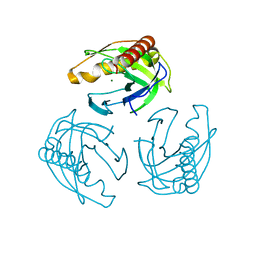 | | Crystal structure of Inorganic Pyrophosphatase PPA1 from Arabidopsis thaliana | | 分子名称: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Soluble inorganic pyrophosphatase 1 | | 著者 | Grzechowiak, M, Sikorski, M, Jaskolski, M. | | 登録日 | 2016-08-22 | | 公開日 | 2017-09-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Crystal structures of plant inorganic pyrophosphatase, an enzyme with a moonlighting autoproteolytic activity.
Biochem.J., 476, 2019
|
|
3KL9
 
 | |
4NJR
 
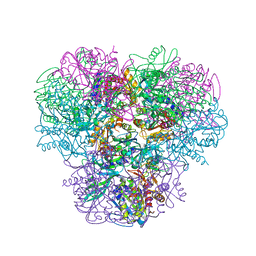 | | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa | | 分子名称: | CARBONATE ION, Probable M18 family aminopeptidase 2, ZINC ION | | 著者 | Nguyen, D.D, Pandian, R, Kim, D.Y, Ha, S.C, Yun, K.H, Kim, K.S, Kim, J.H, Kim, K.K. | | 登録日 | 2013-11-11 | | 公開日 | 2014-04-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa
Biochem.Biophys.Res.Commun., 447, 2014
|
|
4NJQ
 
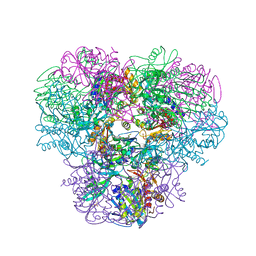 | | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CARBONATE ION, COBALT (II) ION, ... | | 著者 | Nguyen, D.D, Pandian, R, Kim, D.Y, Ha, S.C, Yun, K.H, Kim, K.S, Kim, J.H, Kim, K.K. | | 登録日 | 2013-11-11 | | 公開日 | 2014-04-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.702 Å) | | 主引用文献 | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa
Biochem.Biophys.Res.Commun., 447, 2014
|
|
