2O0O
 
 | | Crystal structure of TL1A | | 分子名称: | MAGNESIUM ION, TNF superfamily ligand TL1A | | 著者 | Jin, T.C, Kim, S, Guo, F, Howard, A.J, Zhang, Y.Z. | | 登録日 | 2006-11-27 | | 公開日 | 2007-10-30 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | X-ray crystal structure of TNF ligand family member TL1A at 2.1A.
Biochem.Biophys.Res.Commun., 364, 2007
|
|
2NML
 
 | | Crystal structure of HEF2/ERH at 1.55 A resolution | | 分子名称: | Enhancer of rudimentary homolog | | 著者 | Jin, T.C, Guo, F, Serebriiskii, I.G, Howard, A.J, Zhang, Y.Z. | | 登録日 | 2006-10-21 | | 公開日 | 2006-10-31 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | A 1.55 A resolution X-ray crystal structure of HEF2/ERH and insights into its transcriptional and cell-cycle interaction networks.
Proteins, 68, 2007
|
|
6JLC
 
 | |
8I6K
 
 | | Structure of hMNDA HIN with dsDNA | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*GP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*C)-3'), ... | | 著者 | Li, Y.L, Jin, T.C. | | 登録日 | 2023-01-28 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural mechanism of dsDNA recognition by the hMNDA HIN domain: New insights into the DNA-binding model of a PYHIN protein.
Int.J.Biol.Macromol., 245, 2023
|
|
7ESV
 
 | |
7ESQ
 
 | |
7ESP
 
 | |
7ESO
 
 | |
5ZGM
 
 | | Crystal Structure of Parvalbumin SPVI, the Major Allergens in Mustelus griseus | | 分子名称: | CALCIUM ION, Parvalbumin SPVI, SULFATE ION | | 著者 | Yang, R.Q, Chen, Y.L, Jin, T.C, Cao, M.J. | | 登録日 | 2018-03-09 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.399 Å) | | 主引用文献 | Purification, Characterization, and Crystal Structure of Parvalbumins, the Major Allergens in Mustelus griseus.
J. Agric. Food Chem., 66, 2018
|
|
5ZH6
 
 | | Crystal structure of Parvalbumin SPV-II of Mustelus griseus | | 分子名称: | CALCIUM ION, Parvalbumin SPV-II, SULFATE ION | | 著者 | Yang, R.Q, Chen, Y.L, Jin, T.C, Cao, M.J. | | 登録日 | 2018-03-11 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Purification, Characterization, and Crystal Structure of Parvalbumins, the Major Allergens in Mustelus griseus.
J. Agric. Food Chem., 66, 2018
|
|
5ZHQ
 
 | |
6AG0
 
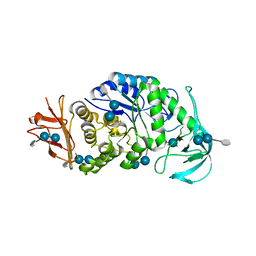 | | The X-ray Crystallographic Structure of Maltooligosaccharide-forming Amylase from Bacillus stearothermophilus STB04 | | 分子名称: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-amylase, CALCIUM ION | | 著者 | Li, Z.F, Li, Y.L, Ban, X.F, Zhang, C.Y, Jin, T.C, Xie, X.F, Gu, Z.B, Li, C.M. | | 登録日 | 2018-08-09 | | 公開日 | 2018-10-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of a maltooligosaccharide-forming amylase from Bacillus stearothermophilus STB04.
Int.J.Biol.Macromol., 138, 2019
|
|
6IWK
 
 | | The Structure of Maltooligosaccharide-forming Amylase from Pseudomonas saccharophila STB07 | | 分子名称: | CALCIUM ION, GLYCEROL, Glucan 1,4-alpha-maltotetraohydrolase | | 著者 | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | 登録日 | 2018-12-05 | | 公開日 | 2019-12-11 | | 最終更新日 | 2021-03-31 | | 実験手法 | X-RAY DIFFRACTION (1.501 Å) | | 主引用文献 | Structure of maltotetraose-forming amylase from Pseudomonas saccharophila STB07 provides insights into its product specificity.
Int.J.Biol.Macromol., 154, 2020
|
|
6KI0
 
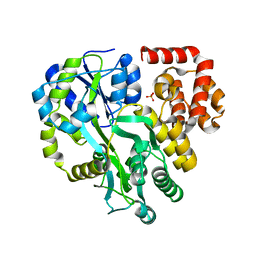 | | Crystal Structure of Human ASC-CARD | | 分子名称: | Maltose/maltodextrin-binding periplasmic protein,Apoptosis-associated speck-like protein containing a CARD, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Xu, Z.H, Jin, T.C. | | 登録日 | 2019-07-16 | | 公開日 | 2020-07-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Homotypic CARD-CARD interaction is critical for the activation of NLRP1 inflammasome.
Cell Death Dis, 12, 2021
|
|
6JQB
 
 | | The structure of maltooligosaccharide-forming amylase from Pseudomonas saccharophila STB07 with pseudo-maltoheptaose | | 分子名称: | 1,2-ETHANEDIOL, ACARBOSE DERIVED HEPTASACCHARIDE, CALCIUM ION, ... | | 著者 | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | 登録日 | 2019-03-30 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.101 Å) | | 主引用文献 | Structure of maltotetraose-forming amylase from Pseudomonas saccharophila STB07 provides insights into its product specificity.
Int.J.Biol.Macromol., 154, 2020
|
|
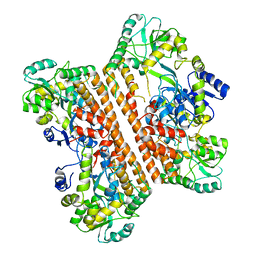
6KXG
 
 | |
7V4Q
 
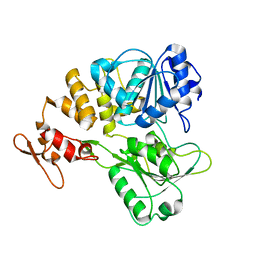 | |
7VOA
 
 | |
7X4I
 
 | |
7V4R
 
 | |
6JOY
 
 | | The X-ray Crystallographic Structure of Branching Enzyme from Rhodothermus obamensis STB05 | | 分子名称: | 1,4-alpha-glucan branching enzyme GlgB | | 著者 | Li, Z.F, Ban, X.F, Jiang, H.M, Wang, Z, Jin, T.C, Li, C.M, Gu, Z.B. | | 登録日 | 2019-03-25 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.392 Å) | | 主引用文献 | Flexible Loop in Carbohydrate-Binding Module 48 Allosterically Modulates Substrate Binding of the 1,4-alpha-Glucan Branching Enzyme.
J.Agric.Food Chem., 69, 2021
|
|
