4A0L
 
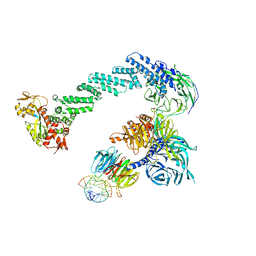 | | Structure of DDB1-DDB2-CUL4B-RBX1 bound to a 12 bp abasic site containing DNA-duplex | | 分子名称: | 12 BP DNA DUPLEX, 12 BP THF CONTAINING DNA DUPLEX, CULLIN-4B, ... | | 著者 | Fischer, E.S, Scrima, A, Gut, H, Thoma, N.H. | | 登録日 | 2011-09-09 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (7.4 Å) | | 主引用文献 | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
6R90
 
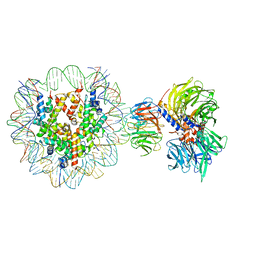 | | Cryo-EM structure of NCP-THF2(+1)-UV-DDB class A | | 分子名称: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R91
 
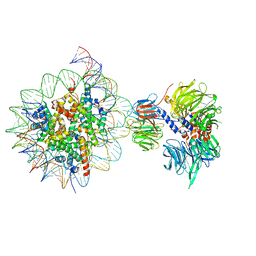 | | Cryo-EM structure of NCP_THF2(-3)-UV-DDB | | 分子名称: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R92
 
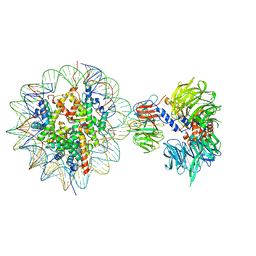 | | Cryo-EM structure of NCP-THF2(+1)-UV-DDB class B | | 分子名称: | DNA damage-binding protein 1,DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R8Z
 
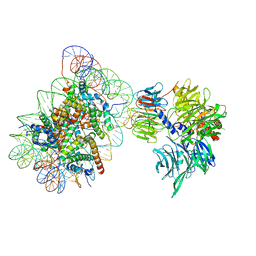 | | Cryo-EM structure of NCP_THF2(-1)-UV-DDB | | 分子名称: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R94
 
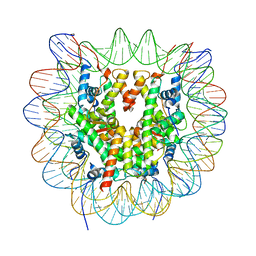 | | Cryo-EM structure of NCP_THF2(-3) | | 分子名称: | Histone H2A type 1-B/E, Histone H2B type 1-J, Histone H3.1, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R8Y
 
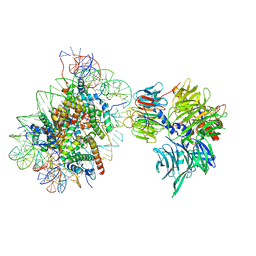 | | Cryo-EM structure of NCP-6-4PP(-1)-UV-DDB | | 分子名称: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R93
 
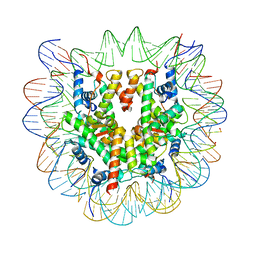 | | Cryo-EM structure of NCP-6-4PP | | 分子名称: | Histone H2A type 1-B/E, Histone H2B type 1-J, Histone H3.1, ... | | 著者 | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | 登録日 | 2019-04-02 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
3TC3
 
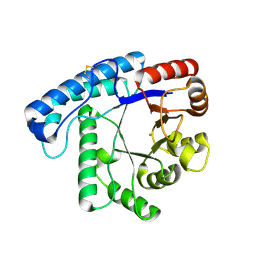 | | Crystal Structure of SacUVDE | | 分子名称: | MANGANESE (II) ION, UV damage endonuclease | | 著者 | Meulenbroek, E.M, Jala, I, Moolenaar, G.F, Goosen, N, Pannu, N.S. | | 登録日 | 2011-08-08 | | 公開日 | 2012-11-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | UV damage endonuclease employs a novel dual-dinucleotide flipping mechanism to recognize different DNA lesions.
Nucleic Acids Res., 41, 2013
|
|
3UMV
 
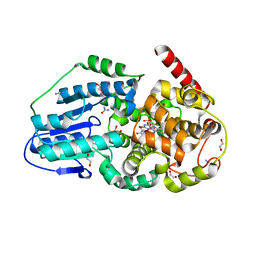 | | Eukaryotic Class II CPD photolyase structure reveals a basis for improved UV-tolerance in plants | | 分子名称: | 1,2-ETHANEDIOL, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Arvai, A.S, Hitomi, K, Getzoff, E.D, Tainer, J.A. | | 登録日 | 2011-11-14 | | 公開日 | 2011-12-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.705 Å) | | 主引用文献 | Eukaryotic Class II Cyclobutane Pyrimidine Dimer Photolyase Structure Reveals Basis for Improved Ultraviolet Tolerance in Plants.
J.Biol.Chem., 287, 2012
|
|
4A0C
 
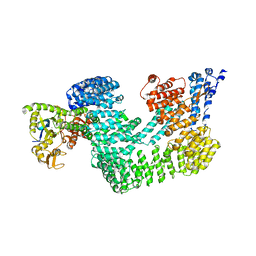 | | Structure of the CAND1-CUL4B-RBX1 complex | | 分子名称: | CULLIN-4B, CULLIN-ASSOCIATED NEDD8-DISSOCIATED PROTEIN 1, E3 UBIQUITIN-PROTEIN LIGASE RBX1, ... | | 著者 | Scrima, A, Fischer, E.S, Faty, M, Gut, H, Thoma, N.H. | | 登録日 | 2011-09-08 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
