6P3Y
 
 | |
6P40
 
 | |
3F88
 
 | | glycogen synthase Kinase 3beta inhibitor complex | | 分子名称: | 3-methylbenzonitrile, 5-[1-(4-methoxyphenyl)-1H-benzimidazol-6-yl]-1,3,4-oxadiazole-2(3H)-thione, Glycogen synthase kinase-3 beta | | 著者 | Mol, C.D, Dougan, D.R. | | 登録日 | 2008-11-11 | | 公開日 | 2009-03-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Design, synthesis and structure-activity relationships of 1,3,4-oxadiazole derivatives as novel inhibitors of glycogen synthase kinase-3beta.
Bioorg.Med.Chem., 17, 2009
|
|
6T9W
 
 | | Crystal structure of formate dehydrogenase FDH2 D222A/Q223R enzyme from Granulicella mallensis MP5ACTX8 in complex with NADP and azide. | | 分子名称: | AZIDE ION, Formate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Robescu, M.S, Rubini, R, Filippini, F, Bergantino, B, Cendron, L. | | 登録日 | 2019-10-29 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | From the Amelioration of a NADP+-dependent Formate Dehydrogenase to the Discovery of a New Enzyme: Round Trip from Theory to Practice
Chemcatchem, 2020
|
|
6TB6
 
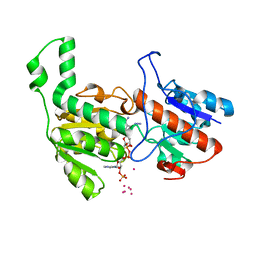 | | Crystal structure of formate dehydrogenase FDH2 D222S/Q223R enzyme from Granulicella mallensis MP5ACTX8 in complex with NADP and azide. | | 分子名称: | AZIDE ION, COBALT (II) ION, Formate dehydrogenase, ... | | 著者 | Robescu, M.S, Rubini, R, Filippini, F, Bergantino, B, Cendron, L. | | 登録日 | 2019-11-01 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | From the Amelioration of a NADP+-dependent Formate Dehydrogenase to the Discovery of a New Enzyme: Round Trip from Theory to Practice
Chemcatchem, 2020
|
|
8BXX
 
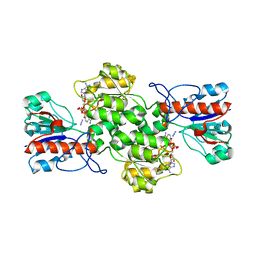 | | Crystal structure of formate dehydrogenase FDH2 enzyme from Granulicella mallensis MP5ACTX8 in complex with NAD and azide. | | 分子名称: | 1,2-ETHANEDIOL, AZIDE ION, Formate dehydrogenase, ... | | 著者 | Robescu, M.S, Rubini, R, Filippini, F, Bergantino, B, Cendron, L. | | 登録日 | 2022-12-10 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | From the amelioration of a NADP+-dependent formate dehydrogenase to the discovery of a new enzyme: round trip from theory to practice
ChemCatChem, 2020
|
|
3GB2
 
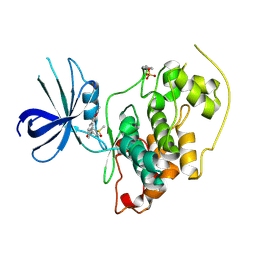 | | GSK3beta inhibitor complex | | 分子名称: | 2-methyl-5-(3-{4-[(S)-methylsulfinyl]phenyl}-1-benzofuran-5-yl)-1,3,4-oxadiazole, Glycogen synthase kinase-3 beta | | 著者 | Mol, C.D. | | 登録日 | 2009-02-18 | | 公開日 | 2010-03-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | 2-{3-[4-(Alkylsulfinyl)phenyl]-1-benzofuran-5-yl}-5-methyl-1,3,4-oxadiazole derivatives as novel inhibitors of glycogen synthase kinase-3beta with good brain permeability.
J.Med.Chem., 52, 2009
|
|
3F7Z
 
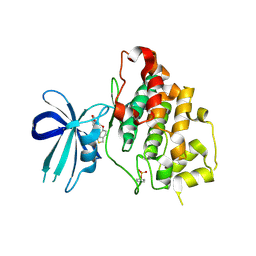 | | X-ray Co-Crystal Structure of Glycogen Synthase Kinase 3beta in Complex with an Inhibitor | | 分子名称: | 2-(1,3-benzodioxol-5-yl)-5-[(3-fluoro-4-methoxybenzyl)sulfanyl]-1,3,4-oxadiazole, Glycogen synthase kinase-3 beta | | 著者 | Mol, C.D, Dougan, D.R. | | 登録日 | 2008-11-10 | | 公開日 | 2009-03-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Design, synthesis and structure-activity relationships of 1,3,4-oxadiazole derivatives as novel inhibitors of glycogen synthase kinase-3beta.
Bioorg.Med.Chem., 17, 2009
|
|
6T9X
 
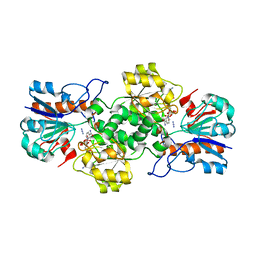 | | Crystal structure of formate dehydrogenase FDH2 D222Q/Q223R mutant enzyme from Granulicella mallensis MP5ACTX8 in complex with NADP and Azide. | | 分子名称: | AZIDE ION, Formate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Robescu, M.S, Rubini, R, Filippini, F, Bergantino, B, Cendron, L. | | 登録日 | 2019-10-29 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | From the Amelioration of a NADP+-dependent Formate Dehydrogenase to the Discovery of a New Enzyme: Round Trip from Theory to Practice
Chemcatchem, 2020
|
|
6T8C
 
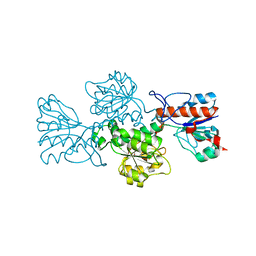 | | Crystal structure of formate dehydrogenase FDH2 enzyme from Granulicella mallensis MP5ACTX8 in the apo form. | | 分子名称: | Formate dehydrogenase | | 著者 | Robescu, M.S, Rubini, R, Filippini, F, Bergantino, B, Cendron, L. | | 登録日 | 2019-10-24 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | From the Amelioration of a NADP+-dependent Formate Dehydrogenase to the Discovery of a New Enzyme: Round Trip from Theory to Practice
Chemcatchem, 2020
|
|
7B3E
 
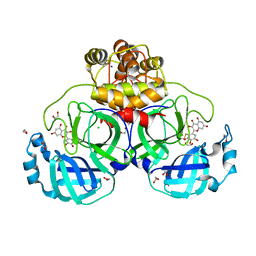 | | Crystal structure of myricetin covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, CHLORIDE ION, ... | | 著者 | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | 登録日 | 2020-11-30 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Identification of Inhibitors of SARS-CoV-2 3CL-Pro Enzymatic Activity Using a Small Molecule in Vitro Repurposing Screen.
Acs Pharmacol Transl Sci, 4, 2021
|
|
1Y2S
 
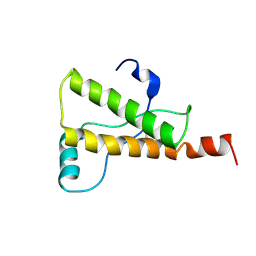 | | Ovine Prion Protein Variant R168 | | 分子名称: | Major prion protein | | 著者 | Christen, B, Lysek, D.A, Herrmann, T, Wuthrich, K. | | 登録日 | 2004-11-23 | | 公開日 | 2004-12-28 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Prion protein NMR structures of cats, dogs, pigs, and sheep
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1XYU
 
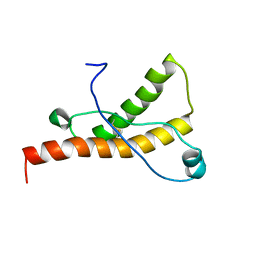 | | Solution structure of the sheep prion protein with polymorphism H168 | | 分子名称: | Major prion protein | | 著者 | Calzolai, L, Lysek, D.A, Guntert, P, Wuthrich, K. | | 登録日 | 2004-11-11 | | 公開日 | 2005-01-04 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Prion protein NMR structures of cats, dogs, pigs, and sheep
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1XFR
 
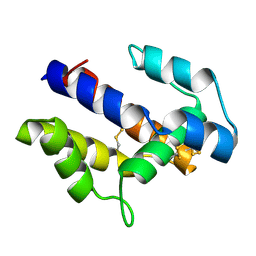 | |
1XYK
 
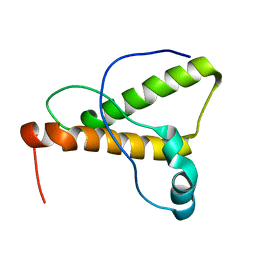 | | NMR Structure of the canine prion protein | | 分子名称: | prion protein | | 著者 | Lysek, D.A, Schorn, C, Esteve-Moya, V, Herrmann, T, Wuthrich, K. | | 登録日 | 2004-11-10 | | 公開日 | 2005-01-04 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Prion protein NMR structures of cats, dogs, pigs, and sheep
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1XYQ
 
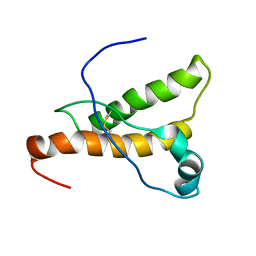 | | NMR structure of the pig prion protein | | 分子名称: | Major prion protein | | 著者 | Lysek, D.A, Schorn, C, Herrmann, T, Wuthrich, K. | | 登録日 | 2004-11-10 | | 公開日 | 2005-01-04 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Prion protein NMR structures of cats, dogs, pigs, and sheep
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
7Q5F
 
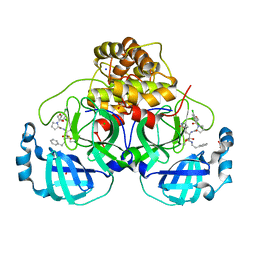 | | Crystal structure of F2F-2020216-01X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | 分子名称: | (S)-1-(2-(2,4-dichlorophenoxy)acetyl)-N-((S)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)-4-(phenethylamino)butan-2-yl)pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, 3C-like proteinase, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2021-11-03 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Easy access to alpha-ketoamides as SARS-CoV-2 and MERS M pro inhibitors via the PADAM oxidation route.
Eur.J.Med.Chem., 244, 2022
|
|
7Q5E
 
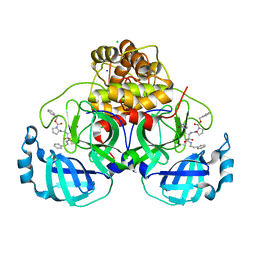 | | Crystal structure of F2F-2020209-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | 分子名称: | 3C-like proteinase, CHLORIDE ION, SODIUM ION, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2021-11-03 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Easy access to alpha-ketoamides as SARS-CoV-2 and MERS M pro inhibitors via the PADAM oxidation route.
Eur.J.Med.Chem., 244, 2022
|
|
1Q90
 
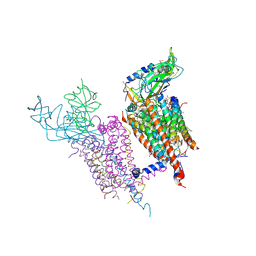 | | Structure of the cytochrome b6f (plastohydroquinone : plastocyanin oxidoreductase) from Chlamydomonas reinhardtii | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 8-HYDROXY-5,7-DIMETHOXY-3-METHYL-2-TRIDECYL-4H-CHROMEN-4-ONE, ... | | 著者 | Stroebel, D, Choquet, Y, Popot, J.-L, Picot, D. | | 登録日 | 2003-08-22 | | 公開日 | 2003-12-09 | | 最終更新日 | 2021-03-03 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | An Atypical Haem in the Cytochrome B6F Complex
Nature, 426, 2003
|
|
7NG3
 
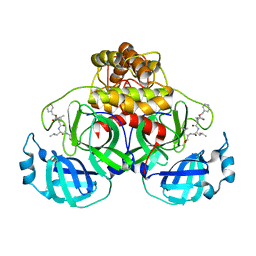 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup P1. | | 分子名称: | 3C-like proteinase, CHLORIDE ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | | 著者 | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | 登録日 | 2021-02-08 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7NG6
 
 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup P1 in absence of DTT. | | 分子名称: | 3C-like proteinase, ACETATE ION, CHLORIDE ION, ... | | 著者 | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | 登録日 | 2021-02-08 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7NF5
 
 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup C2. | | 分子名称: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | 登録日 | 2021-02-05 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7ALI
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 at 1.65A resolution (spacegroup P2(1)). | | 分子名称: | 3C-like proteinase | | 著者 | Costanzi, E, Demitri, N, Giabbai, B, Heroux, A, Storici, P. | | 登録日 | 2020-10-06 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7ALH
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 at 1.65A resolution (spacegroup C2). | | 分子名称: | 3C-like proteinase | | 著者 | Costanzi, E, Demitri, N, Giabbai, B, Heroux, A, Storici, P. | | 登録日 | 2020-10-06 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
2JMR
 
 | | NMR structure of the E. coli type 1 pilus subunit FimF | | 分子名称: | fimF | | 著者 | Gossert, A.D, Bettendorff, P, Puorger, C, Vetsch, M, Herrmann, T, Fiorito, F, Hiller, S, Glockshuber, R, Wuthrich, K. | | 登録日 | 2006-11-29 | | 公開日 | 2007-10-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the Escherichia coli type 1 pilus subunit FimF and its interactions with other pilus subunits.
J.Mol.Biol., 375, 2008
|
|
