7FGK
 
 | | The Fab antibody single structure against tau protein. | | 分子名称: | Fab Heavy Chain, Fab Light Chain, GLYCEROL, ... | | 著者 | Tsuchida, T, Susa, K, Kibiki, T, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, T, Ishida, T, Tomoo, K. | | 登録日 | 2021-07-27 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The free structure of the Fab domain of antibody that recognizes the PHF core region VQIINK in Tau protein.
To Be Published
|
|
7FGJ
 
 | | The cross-reaction complex structure with VQILNK peptide and the tau antibody's Fab domain. | | 分子名称: | CHLORIDE ION, Fab Heavy Chain, Fab Light Chain, ... | | 著者 | Tsuchida, T, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, T, Ishida, T, Tomoo, K. | | 登録日 | 2021-07-27 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | The cross-reaction complex structure with VQILNK peptide and the antibody's Fab domain.
To Be Published
|
|
7FGR
 
 | | The cross-reaction complex structure with VQIFNK peptide and the tau antibody's Fab domain. | | 分子名称: | AMMONIUM ION, CHLORIDE ION, Fab Heavy Chain, ... | | 著者 | Tsuchida, T, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, T, Ishida, T, Tomoo, K. | | 登録日 | 2021-07-27 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The cross-reaction complex structure with VQIFNK peptide and the tau antibody's Fab domain.
To Be Published
|
|
1VCL
 
 | | Crystal Structure of Hemolytic Lectin CEL-III | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Uchida, T, Yamasaki, T, Eto, S, Sugawara, H, Kurisu, G, Nakagawa, A, Kusunoki, M, Hatakeyama, T. | | 登録日 | 2004-03-09 | | 公開日 | 2004-09-07 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal Structure of the Hemolytic Lectin CEL-III Isolated from the Marine Invertebrate Cucumaria echinata: IMPLICATIONS OF DOMAIN STRUCTURE FOR ITS MEMBRANE PORE-FORMATION MECHANISM
J.Biol.Chem., 279, 2004
|
|
3GH7
 
 | | Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with GalNAc | | 分子名称: | 2-acetamido-2-deoxy-beta-D-galactopyranose, SULFATE ION, beta-hexosaminidase | | 著者 | Sumida, T, Ishii, R, Yanagisawa, T, Yokoyama, S, Ito, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2009-03-03 | | 公開日 | 2009-07-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Molecular cloning and crystal structural analysis of a novel beta-N-acetylhexosaminidase from Paenibacillus sp. TS12 capable of degrading glycosphingolipids
J.Mol.Biol., 392, 2009
|
|
3GH5
 
 | | Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with GlcNAc | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, beta-hexosaminidase | | 著者 | Sumida, T, Ishii, R, Yanagisawa, T, Yokoyama, S, Ito, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2009-03-03 | | 公開日 | 2009-07-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Molecular cloning and crystal structural analysis of a novel beta-N-acetylhexosaminidase from Paenibacillus sp. TS12 capable of degrading glycosphingolipids
J.Mol.Biol., 392, 2009
|
|
3GH4
 
 | | Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 | | 分子名称: | ACETIC ACID, SULFATE ION, beta-hexosaminidase | | 著者 | Sumida, T, Ishii, R, Yanagisawa, T, Yokoyama, S, Ito, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2009-03-03 | | 公開日 | 2009-07-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Molecular cloning and crystal structural analysis of a novel beta-N-acetylhexosaminidase from Paenibacillus sp. TS12 capable of degrading glycosphingolipids
J.Mol.Biol., 392, 2009
|
|
3EQN
 
 | | Crystal structure of beta-1,3-glucanase from Phanerochaete chrysosporium (Lam55A) | | 分子名称: | ACETATE ION, GLYCEROL, Glucan 1,3-beta-glucosidase, ... | | 著者 | Ishida, T, Fushinobu, S, Kawai, R, Kitaoka, M, Igarashi, K, Samejima, M. | | 登録日 | 2008-10-01 | | 公開日 | 2009-02-03 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of glycoside hydrolase family 55 beta -1,3-glucanase from the basidiomycete Phanerochaete chrysosporium
J.Biol.Chem., 284, 2009
|
|
3EQO
 
 | | Crystal structure of beta-1,3-glucanase from Phanerochaete chrysosporium (Lam55A) gluconolactone complex | | 分子名称: | D-glucono-1,5-lactone, Glucan 1,3-beta-glucosidase, ZINC ION, ... | | 著者 | Ishida, T, Fushinobu, S, Kawai, R, Kitaoka, M, Igarashi, K, Samejima, M. | | 登録日 | 2008-10-01 | | 公開日 | 2009-02-03 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal structure of glycoside hydrolase family 55 beta -1,3-glucanase from the basidiomycete Phanerochaete chrysosporium
J.Biol.Chem., 284, 2009
|
|
3MM2
 
 | | Dye-decolorizing peroxidase (DyP) in complex with cyanide | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYANIDE ION, DyP, ... | | 著者 | Sugano, Y, Yoshida, T, Tsuge, H. | | 登録日 | 2010-04-19 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The catalytic mechanism of dye-decolorizing peroxidase DyP may require the swinging movement of an aspartic acid residue
Febs J., 278, 2011
|
|
7CEG
 
 | | Crystal structure of the complex between mouse PTP delta and neuroligin-3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform C of Receptor-type tyrosine-protein phosphatase delta, Neuroligin-3 | | 著者 | Yamagata, A, Yoshida, T, Shiroshima, T, Maeda, A, Fukai, S. | | 登録日 | 2020-06-23 | | 公開日 | 2021-02-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.85 Å) | | 主引用文献 | Canonical versus non-canonical transsynaptic signaling of neuroligin 3 tunes development of sociality in mice.
Nat Commun, 12, 2021
|
|
7CEE
 
 | | Crystal structure of mouse neuroligin-3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neuroligin-3 | | 著者 | Yamagata, A, Yoshida, T, Shiroshima, T, Maeda, A, Fukai, S. | | 登録日 | 2020-06-23 | | 公開日 | 2021-02-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.763 Å) | | 主引用文献 | Canonical versus non-canonical transsynaptic signaling of neuroligin 3 tunes development of sociality in mice.
Nat Commun, 12, 2021
|
|
8JF5
 
 | | Crystal structure of Lysine Specific Demethylase 1 (LSD1) with TAS1440 | | 分子名称: | 4-[5-[(3~{R})-3-azanylpyrrolidin-1-yl]carbonyl-2-[2-fluoranyl-4-(2-methyl-2-oxidanyl-propyl)phenyl]phenyl]-2-fluoranyl-benzenecarbonitrile, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | 著者 | Fukushima, H, Machida, T, Yamashita, S, Suzuki, T. | | 登録日 | 2023-05-17 | | 公開日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | TAS1440, a histone H3 competitive LSD1 inhibitor, activates both TGF-beta and notch signaling pathways via INSM1 dissociation in neuroendocrine small cell lung cancer
To Be Published
|
|
3MM3
 
 | |
3MM1
 
 | | Dye-decolorizing peroxidase (DyP) D171N | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DyP, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Sugano, Y, Yoshida, T, Tsuge, H. | | 登録日 | 2010-04-19 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | The catalytic mechanism of dye-decolorizing peroxidase DyP may require the swinging movement of an aspartic acid residue
Febs J., 278, 2011
|
|
6KLO
 
 | | Complex structure of Iota toxin enzymatic component (Ia) and binding component (Ib) pore with short stem | | 分子名称: | CALCIUM ION, Iota toxin component Ia, Iota toxin component Ib | | 著者 | Yoshida, T, Yamada, T, Kawamoto, A, Mitsuoka, K, Iwasaki, K, Tsuge, H. | | 登録日 | 2019-07-30 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6K9Z
 
 | | STRUCTURE OF URIDYLYLTRANSFERASE MUTANT | | 分子名称: | ACETATE ION, FE (III) ION, Galactose-1-phosphate uridylyltransferase, ... | | 著者 | Sakuraba, H, Ohshida, T, Yoneda, K, Ohshima, T. | | 登録日 | 2019-06-19 | | 公開日 | 2019-12-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Unique active site formation in a novel galactose 1-phosphate uridylyltransferase from the hyperthermophilic archaeon Pyrobaculum aerophilum.
Proteins, 88, 2020
|
|
6KLW
 
 | | Complex structure of Iota toxin enzymatic component (Ia) and binding component (Ib) pore with long stem | | 分子名称: | CALCIUM ION, Iota toxin component Ia, Iota toxin component Ib | | 著者 | Yoshida, T, Yamada, T, Kawamoto, A, Mitsuoka, K, Iwasaki, K, Tsuge, H. | | 登録日 | 2019-07-30 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6KLX
 
 | | Pore structure of Iota toxin binding component (Ib) | | 分子名称: | CALCIUM ION, Iota toxin component Ib | | 著者 | Yoshida, T, Yamada, T, Kawamoto, A, Mitsuoka, K, Iwasaki, K, Tsuge, H. | | 登録日 | 2019-07-30 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6K5Z
 
 | | Structure of uridylyltransferase | | 分子名称: | FE (III) ION, Galactose-1-phosphate uridylyltransferase, PHOSPHATE ION, ... | | 著者 | Sakuraba, H, Ohshida, T, Yoneda, K, Ohshima, T. | | 登録日 | 2019-05-31 | | 公開日 | 2019-12-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Unique active site formation in a novel galactose 1-phosphate uridylyltransferase from the hyperthermophilic archaeon Pyrobaculum aerophilum.
Proteins, 88, 2020
|
|
1PPP
 
 | |
3VJK
 
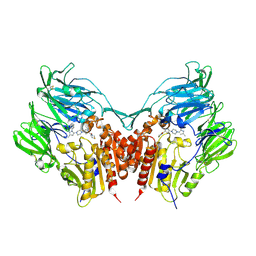 | | Crystal structure of human depiptidyl peptidase IV (DPP-4) in complex with MP-513 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | 著者 | Akahoshi, F, Kishida, H, Miyaguchi, I, Yoshida, T, Ishii, S. | | 登録日 | 2011-10-24 | | 公開日 | 2012-10-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Discovery and preclinical profile of teneligliptin (3-[(2S,4S)-4-[4-(3-methyl-1-phenyl-1H-pyrazol-5-yl)piperazin-1-yl]pyrrolidin-2-ylcarbonyl]thiazolidine): A highly potent, selective, long-lasting and orally active dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes
Bioorg.Med.Chem., 20, 2012
|
|
3VJL
 
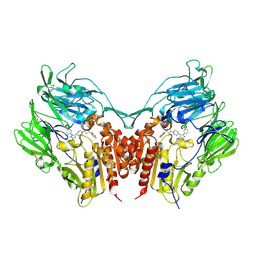 | | Crystal structure of human depiptidyl peptidase IV (DPP-4) in complex with a prolylthiazolidine inhibitor #2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | 著者 | Akahoshi, F, Kishida, H, Miyaguchi, I, Yoshida, T, Ishii, S. | | 登録日 | 2011-10-24 | | 公開日 | 2012-10-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.393 Å) | | 主引用文献 | Discovery and preclinical profile of teneligliptin (3-[(2S,4S)-4-[4-(3-methyl-1-phenyl-1H-pyrazol-5-yl)piperazin-1-yl]pyrrolidin-2-ylcarbonyl]thiazolidine): A highly potent, selective, long-lasting and orally active dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes
Bioorg.Med.Chem., 20, 2012
|
|
3A5Z
 
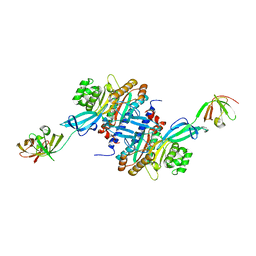 | | Crystal structure of Escherichia coli GenX in complex with elongation factor P | | 分子名称: | 5'-O-[(L-LYSYLAMINO)SULFONYL]ADENOSINE, Elongation factor P, Putative lysyl-tRNA synthetase | | 著者 | Sumida, T, Yanagisawa, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2009-08-17 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A paralog of lysyl-tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3A5Y
 
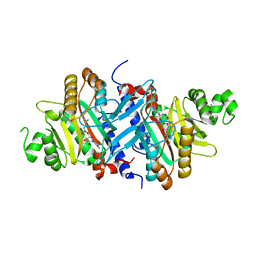 | | Crystal structure of GenX from Escherichia coli in complex with lysyladenylate analog | | 分子名称: | 5'-O-[(L-LYSYLAMINO)SULFONYL]ADENOSINE, Putative lysyl-tRNA synthetase | | 著者 | Sumida, T, Yanagisawa, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2009-08-17 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A paralog of lysyl-tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P.
Nat.Struct.Mol.Biol., 17, 2010
|
|
