1M11
 
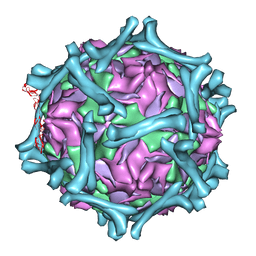 | | structural model of human decay-accelerating factor bound to echovirus 7 from cryo-electron microscopy | | 分子名称: | COAT PROTEIN VP1, COAT PROTEIN VP2, COAT PROTEIN VP3, ... | | 著者 | He, Y, Lin, F, Chipman, P.R, Bator, C.M, Baker, T.S, Shoham, M, Kuhn, R.J, Medof, M.E, Rossmann, M.G. | | 登録日 | 2002-06-17 | | 公開日 | 2002-08-28 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (16 Å) | | 主引用文献 | Structure of decay-accelerating factor bound to echovirus 7: a virus-receptor complex.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2KQ8
 
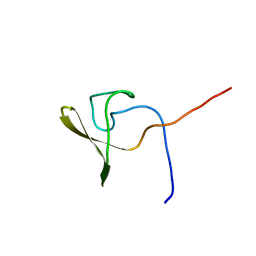 | | Solution NMR structure of a domain from BT9727_4915 from Bacillus thuringiensis, Northeast Structural Genomics Consortium Target BuR95A | | 分子名称: | Cell wall hydrolase | | 著者 | He, Y, Mills, J.L, Wu, Y, Eletsky, A, Wang, H, Ciccosanti, C, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2009-10-30 | | 公開日 | 2010-01-26 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of a domain from BT9727_4915 from Bacillus thuringiensis, Northeast Structural Genomics Consortium Target BuR95A
To be Published
|
|
2LHC
 
 | | Ga98 solution structure | | 分子名称: | Ga98 | | 著者 | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | 登録日 | 2011-08-08 | | 公開日 | 2012-02-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Mutational tipping points for switching protein folds and functions.
Structure, 20, 2012
|
|
2JWS
 
 | | Solution NMR structures of two designed proteins with 88% sequence identity but different fold and function | | 分子名称: | Ga88 | | 著者 | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | 登録日 | 2007-10-24 | | 公開日 | 2008-09-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structures of two designed proteins with high sequence identity but different fold and function
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2JU7
 
 | | Solution-State Structures of Oleate-Liganded LFABP, Protein Only | | 分子名称: | Fatty acid-binding protein, liver | | 著者 | He, Y, Yang, X, Wang, H, Estephan, R, Francis, F, Kodukula, S, Storch, J, Stark, R.E. | | 登録日 | 2007-08-15 | | 公開日 | 2007-11-20 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution-State Molecular Structure of Apo and Oleate-Liganded Liver Fatty Acid-Binding Protein
Biochemistry, 46, 2007
|
|
2LHG
 
 | | GB98-T25I solution structure | | 分子名称: | GB98 | | 著者 | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | 登録日 | 2011-08-08 | | 公開日 | 2012-02-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Mutational tipping points for switching protein folds and functions.
Structure, 20, 2012
|
|
2JWU
 
 | | Solution NMR structures of two designed proteins with 88% sequence identity but different fold and function | | 分子名称: | Gb88 | | 著者 | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | 登録日 | 2007-10-25 | | 公開日 | 2008-09-09 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structures of two designed proteins with high sequence identity but different fold and function
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2JU3
 
 | | Solution-state NMR structures of apo-LFABP (Liver Fatty Acid-Binding Protein) | | 分子名称: | Fatty acid-binding protein, liver | | 著者 | He, Y, Yang, X, Wang, H, Estephan, R, Francis, F, Kodukula, S, Storch, J, Stark, R.E. | | 登録日 | 2007-08-14 | | 公開日 | 2007-11-20 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution-State Molecular Structure of Apo and Oleate-Liganded Liver Fatty Acid-Binding Protein
Biochemistry, 46, 2007
|
|
2LHE
 
 | | Gb98-T25I,L20A | | 分子名称: | Gb98 | | 著者 | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | 登録日 | 2011-08-08 | | 公開日 | 2012-02-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Mutational tipping points for switching protein folds and functions.
Structure, 20, 2012
|
|
2LHD
 
 | | GB98 solution structure | | 分子名称: | GB98 | | 著者 | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | 登録日 | 2011-08-08 | | 公開日 | 2012-02-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Mutational tipping points for switching protein folds and functions.
Structure, 20, 2012
|
|
2LU1
 
 | | pfsub2 solution NMR structure | | 分子名称: | Subtilase | | 著者 | He, Y, Chen, Y, Ruan, B, O'Brochta, D, Bryan, P, Orban, J. | | 登録日 | 2012-06-06 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of a sheddase inhibitor prodomain from the malarial parasite Plasmodium falciparum.
Proteins, 80, 2012
|
|
7YDJ
 
 | | Cryo EM structure of CD97/miniG12 complex | | 分子名称: | Adhesion G protein-coupled receptor E5 subunit beta, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | He, Y, Wang, N. | | 登録日 | 2022-07-04 | | 公開日 | 2023-07-12 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | Cryo EM structure of CD97/miniG12 complex
To Be Published
|
|
4OIC
 
 | | Crystal structrual of a soluble protein | | 分子名称: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Bet v I allergen-like, CHLORIDE ION, ... | | 著者 | He, Y, Hao, Q, Li, W, Yan, C, Yan, N, Yin, P. | | 登録日 | 2014-01-19 | | 公開日 | 2015-01-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.999 Å) | | 主引用文献 | Identification and characterization of ABA receptors in Oryza sativa
Plos One, 9, 2014
|
|
3R8W
 
 | | Structure of 3-isopropylmalate dehydrogenase isoform 2 from Arabidopsis thaliana at 2.2 angstrom resolution | | 分子名称: | 3-isopropylmalate dehydrogenase 2, chloroplastic, ACETATE ION | | 著者 | He, Y, Galant, A, Pang, Q, Strul, J.M, Balogun, S, Jez, J.M, Chen, S. | | 登録日 | 2011-03-24 | | 公開日 | 2011-06-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural and functional evolution of isopropylmalate dehydrogenases in the leucine and glucosinolate pathways of Arabidopsis thaliana.
J.Biol.Chem., 286, 2011
|
|
4P6X
 
 | | Crystal Structure of cortisol-bound glucocorticoid receptor ligand binding domain | | 分子名称: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, Glucocorticoid receptor, Nuclear receptor coactivator 2 | | 著者 | He, Y, Zhou, X.E, Tolbert, W.D, Powell, K, Melcher, K, Xu, H.E. | | 登録日 | 2014-03-25 | | 公開日 | 2014-04-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures and mechanism for the design of highly potent glucocorticoids.
Cell Res., 24, 2014
|
|
4P6W
 
 | | Crystal Structure of mometasone furoate-bound glucocorticoid receptor ligand binding domain | | 分子名称: | Glucocorticoid receptor, MOMETASONE FUROATE, Nuclear receptor coactivator 2 | | 著者 | He, Y, Zhou, X.E, Tolbert, W.D, Powell, K, Melcher, K, Xu, H.E. | | 登録日 | 2014-03-25 | | 公開日 | 2014-04-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.951 Å) | | 主引用文献 | Structures and mechanism for the design of highly potent glucocorticoids.
Cell Res., 24, 2014
|
|
5I8Q
 
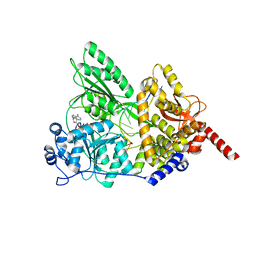 | | S. cerevisiae Prp43 in complex with RNA and ADPNP | | 分子名称: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP43, ... | | 著者 | He, Y, Nielsen, K.H, Andersen, G.R. | | 登録日 | 2016-02-19 | | 公開日 | 2017-02-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (4.2 Å) | | 主引用文献 | Structure of the DEAH/RHA ATPase Prp43p bound to RNA implicates a pair of hairpins and motif Va in translocation along RNA.
RNA, 23, 2017
|
|
5IYA
 
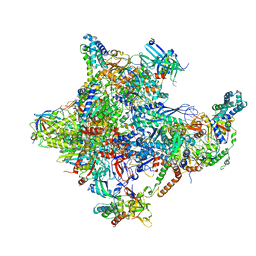 | | Human core-PIC in the closed state | | 分子名称: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB10, DNA-directed RNA polymerase II subunit RPB11-a, ... | | 著者 | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | 登録日 | 2016-03-24 | | 公開日 | 2016-05-18 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (5.4 Å) | | 主引用文献 | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
5IY8
 
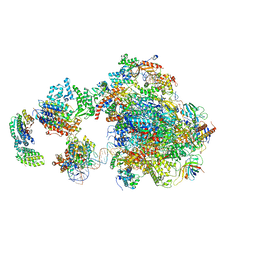 | | Human holo-PIC in the initial transcribing state | | 分子名称: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB10, DNA-directed RNA polymerase II subunit RPB11-a, ... | | 著者 | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | 登録日 | 2016-03-24 | | 公開日 | 2016-05-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (7.9 Å) | | 主引用文献 | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
5IY6
 
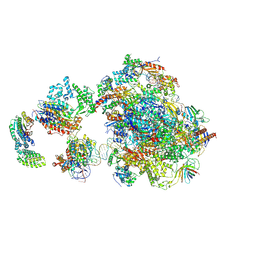 | | Human holo-PIC in the closed state | | 分子名称: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB10, DNA-directed RNA polymerase II subunit RPB11-a, ... | | 著者 | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | 登録日 | 2016-03-24 | | 公開日 | 2016-05-18 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (7.2 Å) | | 主引用文献 | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
5IY9
 
 | | Human holo-PIC in the initial transcribing state (no IIS) | | 分子名称: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB10, DNA-directed RNA polymerase II subunit RPB11-a, ... | | 著者 | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | 登録日 | 2016-03-24 | | 公開日 | 2016-05-18 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (6.3 Å) | | 主引用文献 | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
5IYD
 
 | | Human core-PIC in the initial transcribing state (no IIS) | | 分子名称: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB10, DNA-directed RNA polymerase II subunit RPB11-a, ... | | 著者 | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | 登録日 | 2016-03-24 | | 公開日 | 2016-05-18 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
5IYC
 
 | | Human core-PIC in the initial transcribing state | | 分子名称: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB10, DNA-directed RNA polymerase II subunit RPB11-a, ... | | 著者 | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | 登録日 | 2016-03-24 | | 公開日 | 2016-05-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
5IYB
 
 | | Human core-PIC in the open state | | 分子名称: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB10, DNA-directed RNA polymerase II subunit RPB11-a, ... | | 著者 | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | 登録日 | 2016-03-24 | | 公開日 | 2016-05-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
5IVW
 
 | | Human core TFIIH bound to DNA within the PIC | | 分子名称: | General transcription factor IIH subunit 2, General transcription factor IIH subunit 3, General transcription factor IIH subunit 4, ... | | 著者 | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | 登録日 | 2016-03-21 | | 公開日 | 2016-05-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (10 Å) | | 主引用文献 | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
