8XVY
 
 | |
8XVX
 
 | |
8XW1
 
 | |
8XW4
 
 | |
8XRY
 
 | |
8XW0
 
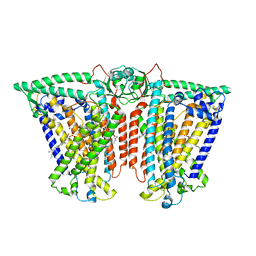 | | Cryo-EM structure of OSCA3.1-GDN state | | 分子名称: | CSC1-like protein ERD4, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine | | 著者 | Zhang, Y, Han, Y. | | 登録日 | 2024-01-15 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 628, 2024
|
|
8XS5
 
 | |
8XW2
 
 | |
8XW3
 
 | |
8XAJ
 
 | |
6AEI
 
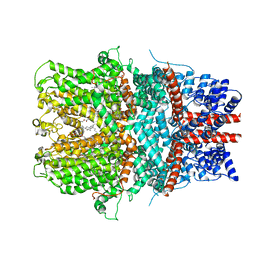 | | Cryo-EM structure of the receptor-activated TRPC5 ion channel | | 分子名称: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | 著者 | Duan, J, Li, Z, Li, J, Zhang, J. | | 登録日 | 2018-08-05 | | 公開日 | 2019-08-07 | | 最終更新日 | 2019-08-14 | | 実験手法 | ELECTRON MICROSCOPY (2.89 Å) | | 主引用文献 | Cryo-EM structure of TRPC5 at 2.8- angstrom resolution reveals unique and conserved structural elements essential for channel function.
Sci Adv, 5, 2019
|
|
6O6W
 
 | | Solution structure of human myeloid-derived growth factor | | 分子名称: | Myeloid-derived growth factor | | 著者 | Bortnov, V, Tonelli, M, Lee, W, Markley, J.L, Mosher, D.F. | | 登録日 | 2019-03-07 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of human myeloid-derived growth factor suggests a conserved function in the endoplasmic reticulum.
Nat Commun, 10, 2019
|
|
6LW5
 
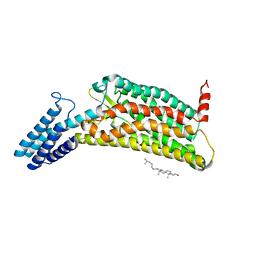 | | Crystal structure of the human formyl peptide receptor 2 in complex with WKYMVm | | 分子名称: | CHOLESTEROL, Soluble cytochrome b562,N-formyl peptide receptor 2, TRP-LYS-TYR-MET-VAL-QXV | | 著者 | Chen, T, Zong, X, Zhang, H, Wang, M, Zhao, Q, Wu, B. | | 登録日 | 2020-02-07 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis of ligand binding modes at the human formyl peptide receptor 2.
Nat Commun, 11, 2020
|
|
7VFX
 
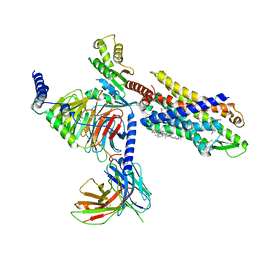 | | The structure of Formyl Peptide Receptor 1 in complex with Gi and peptide agonist fMIFL | | 分子名称: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Wang, X.K, Chen, G, Liao, Q.W, Du, Y, Hu, H.L, Ye, D.Q. | | 登録日 | 2021-09-14 | | 公開日 | 2022-09-21 | | 最終更新日 | 2022-12-07 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural basis for recognition of N-formyl peptides as pathogen-associated molecular patterns.
Nat Commun, 13, 2022
|
|
8WKX
 
 | | Cryo-EM structure of DSR2 | | 分子名称: | SIR2-like domain-containing protein | | 著者 | Gao, A, Huang, J, Zhu, K. | | 登録日 | 2023-09-28 | | 公開日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.15 Å) | | 主引用文献 | Molecular basis of bacterial DSR2 anti-phage defense and viral immune evasion.
Nat Commun, 15, 2024
|
|
8WKS
 
 | |
8WKT
 
 | | Cryo-EM structure of DSR2-DSAD1 complex | | 分子名称: | SIR2-like domain-containing protein, SPbeta prophage-derived uncharacterized protein YotI | | 著者 | Gao, A, Huang, J, Zhu, K. | | 登録日 | 2023-09-28 | | 公開日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.86 Å) | | 主引用文献 | Molecular basis of bacterial DSR2 anti-phage defense and viral immune evasion.
Nat Commun, 15, 2024
|
|
5VIF
 
 | | Electrophilic probes for deciphering substrate recognition by O-GlcNAc transferase | | 分子名称: | 2-{[(2E)-4-chlorobut-2-enoyl]amino}-2-deoxy-beta-D-glucopyranose, CKII, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, ... | | 著者 | Jiang, J, Li, B, Hu, C.-W, Worth, M, Fan, D, Li, H. | | 登録日 | 2017-04-15 | | 公開日 | 2017-10-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Electrophilic probes for deciphering substrate recognition by O-GlcNAc transferase.
Nat. Chem. Biol., 13, 2017
|
|
5VIE
 
 | | Electrophilic probes for deciphering substrate recognition by O-GlcNAc transferase | | 分子名称: | 2-{[(2E)-4-chlorobut-2-enoyl]amino}-2-deoxy-beta-D-glucopyranose, 2-{[(2E)-but-2-enoyl]amino}-2-deoxy-beta-D-glucopyranose, CKII, ... | | 著者 | Jiang, J, Li, B, Hu, C.-W, Worth, M, Fan, D, Li, H. | | 登録日 | 2017-04-15 | | 公開日 | 2017-10-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Electrophilic probes for deciphering substrate recognition by O-GlcNAc transferase.
Nat. Chem. Biol., 13, 2017
|
|
7L3J
 
 | | T4 Lysozyme L99A - benzylacetate - RT | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Fischer, M, Bradford, S.Y.C. | | 登録日 | 2020-12-17 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3A
 
 | |
7L39
 
 | | T4 Lysozyme L99A - toluene - RT | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Fischer, M, Bradford, S.Y.C. | | 登録日 | 2020-12-17 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L38
 
 | | T4 Lysozyme L99A - Apo - cryo | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Fischer, M, Bradford, S.Y.C. | | 登録日 | 2020-12-17 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3B
 
 | | T4 Lysozyme L99A - iodobenzene - RT | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Fischer, M, Bradford, S.Y.C. | | 登録日 | 2020-12-17 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L37
 
 | | T4 Lysozyme L99A - Apo - RT | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Fischer, M, Bradford, S.Y.C. | | 登録日 | 2020-12-17 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.439 Å) | | 主引用文献 | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
