8H51
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (T21I and E166V) in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-11 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H5P
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (L50F and E166V) in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-13 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H6N
 
 | | Crystal structure of SARS-CoV-2 main protease (Mpro) Mutant (T21I) in complex with protease inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 2-(diethylamino)-N-(2,6-dimethylphenyl)ethanamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-18 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H7W
 
 | | Crystal structure of SARS-CoV-2 main protease (Mpro) Mutant (S144A) in complex with protease inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-21 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H57
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) A193P Mutant in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-12 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H82
 
 | | Crystal structure of SARS-CoV-2 main protease (Mpro) Mutant (E166V) in complex with protease inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-21 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8HBK
 
 | | The crystal structure of SARS-CoV-2 3CL protease in complex with Ensitrelvir | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Lin, M. | | 登録日 | 2022-10-29 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H4Y
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) F140L Mutant in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-11 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
7BUY
 
 | | The crystal structure of COVID-19 main protease in complex with carmofur | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, hexylcarbamic acid | | 著者 | Zhao, Y, Zhang, B, Jin, Z, Liu, X, Yang, H, Rao, Z. | | 登録日 | 2020-04-08 | | 公開日 | 2020-04-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6LU7
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor N3 | | 分子名称: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | 著者 | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | 登録日 | 2020-01-26 | | 公開日 | 2020-02-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
8SUT
 
 | | Crystal structure of YisK from Bacillus subtilis in complex with reaction product 4-Hydroxy-2-oxoglutaric acid | | 分子名称: | (2~{R})-2-oxidanyl-4-oxidanylidene-pentanedioic acid, Fumarylacetoacetate hydrolase family protein, MANGANESE (II) ION | | 著者 | Krieger, I.V, Chemelewski, V, Guo, T, Sperber, A, Herman, J, Sacchettini, J.C. | | 登録日 | 2023-05-13 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Bacillus subtilis YisK possesses oxaloacetate decarboxylase activity and exhibits Mbl-dependent localization.
J.Bacteriol., 206, 2024
|
|
8SUU
 
 | | Crystal structure of YisK from Bacillus subtilis in apo form | | 分子名称: | Fumarylacetoacetate hydrolase family protein, MANGANESE (II) ION | | 著者 | Krieger, I.V, Chemelewski, V, Guo, T, Sperber, A, Herman, J, Sacchettini, J.C. | | 登録日 | 2023-05-13 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Bacillus subtilis YisK possesses oxaloacetate decarboxylase activity and exhibits Mbl-dependent localization.
J.Bacteriol., 206, 2024
|
|
8SKY
 
 | | Crystal structure of YisK from Bacillus subtilis in complex with oxalate | | 分子名称: | Fumarylacetoacetate hydrolase family protein, MANGANESE (II) ION, OXALATE ION | | 著者 | Krieger, I.V, Chemelewski, V, Guo, T, Sperber, A, Herman, J, Sacchettini, J.C. | | 登録日 | 2023-04-20 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Bacillus subtilis YisK possesses oxaloacetate decarboxylase activity and exhibits Mbl-dependent localization.
J.Bacteriol., 206, 2024
|
|
5GPR
 
 | | Crystal structure of chitinase-h from Ostrinia furnacalis | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase | | 著者 | Liu, T, Zhou, Y, Chen, L, Yang, Q. | | 登録日 | 2016-08-04 | | 公開日 | 2017-01-11 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.23 Å) | | 主引用文献 | Structure, Catalysis, and Inhibition of OfChi-h, the Lepidoptera-exclusive Insect Chitinase.
J. Biol. Chem., 292, 2017
|
|
5GQB
 
 | | Crystal structure of chitinase-h from O. furnacalis in complex with chitohepatose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Liu, T, Zhou, Y, Chen, L, Yang, Q. | | 登録日 | 2016-08-06 | | 公開日 | 2017-01-11 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure, Catalysis, and Inhibition of OfChi-h, the Lepidoptera-exclusive Insect Chitinase.
J. Biol. Chem., 292, 2017
|
|
1ERI
 
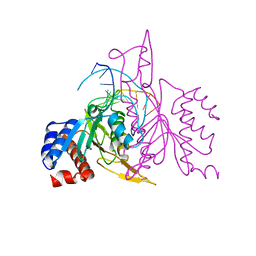 | | X-RAY STRUCTURE OF THE DNA-ECO RI ENDONUCLEASE-DNA RECOGNITION COMPLEX: THE RECOGNITION NETWORK AND THE INTEGRATION OF RECOGNITION AND CLEAVAGE | | 分子名称: | DNA (5'-D(*TP*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), PROTEIN (ECO RI ENDONUCLEASE (E.C.3.1.21.4)) | | 著者 | Kim, Y, Grable, J.C, Love, R, Greene, P.J, Rosenberg, J.M. | | 登録日 | 1994-05-18 | | 公開日 | 1995-02-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing.
Science, 249, 1990
|
|
1XX5
 
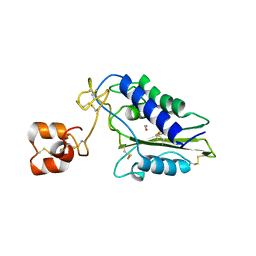 | | Crystal Structure of Natrin from Naja atra snake venom | | 分子名称: | ETHANOL, Natrin 1 | | 著者 | Wang, J, Shen, B, Lou, X.H, Guo, M, Teng, M.K, Niu, L.W. | | 登録日 | 2004-11-04 | | 公開日 | 2005-06-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Blocking effect and crystal structure of natrin toxin, a cysteine-rich secretory protein from Naja atra venom that targets the BKCa channel
Biochemistry, 44, 2005
|
|
8WW2
 
 | | GPR3/Gs complex | | 分子名称: | CHOLESTEROL HEMISUCCINATE, G-protein coupled receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | He, Y, Xiong, Y. | | 登録日 | 2023-10-24 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.79 Å) | | 主引用文献 | Identification of oleic acid as an endogenous ligand of GPR3.
Cell Res., 34, 2024
|
|
8Y7Y
 
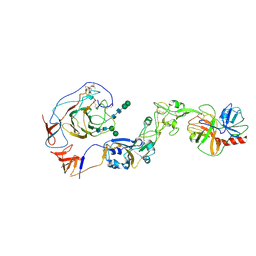 | | Local structure of HCoV-HKU1A spike in complex with TMPRSS2 and glycan | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Wang, H.F, Zhang, X, Lu, Y, Liu, X, Sun, L, Yang, H.T. | | 登録日 | 2024-02-05 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 2024
|
|
8Y8D
 
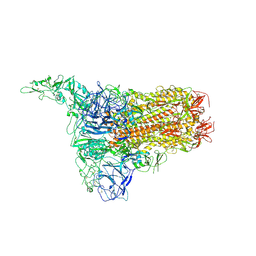 | | Structure of HCoV-HKU1C spike in the inactive-1up conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | 登録日 | 2024-02-06 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.41 Å) | | 主引用文献 | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 2024
|
|
8Y8E
 
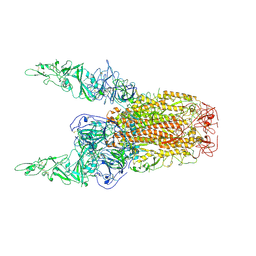 | | Structure of HCoV-HKU1C spike in the inactive-2up conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | 登録日 | 2024-02-06 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.62 Å) | | 主引用文献 | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 2024
|
|
8Y8C
 
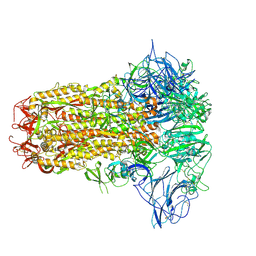 | | Structure of HCoV-HKU1C spike in the inactive-closed conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | 登録日 | 2024-02-06 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 2024
|
|
8Y87
 
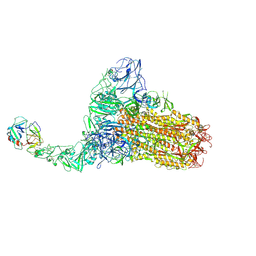 | | Structure of HCoV-HKU1C spike in the functionally anchored-1up conformation with 1TMPRSS2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Lu, Y.C, Zhang, X, Wang, H.F, Liu, X.C, Sun, L, Yang, H.T. | | 登録日 | 2024-02-06 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 2024
|
|
8Y89
 
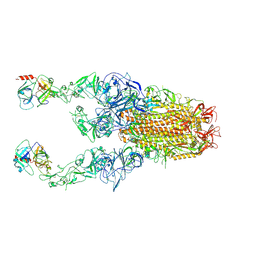 | | Structure of HCoV-HKU1C spike in the functionally anchored-3up conformation with 2TMPRSS2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Lu, Y.C, Wang, H.F, Zhang, X, Liu, X.C, Sun, L, Yang, H.T. | | 登録日 | 2024-02-06 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.32 Å) | | 主引用文献 | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 2024
|
|
8Y8G
 
 | | Structure of HCoV-HKU1C spike in the glycan-activated-1up conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | 登録日 | 2024-02-06 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.23 Å) | | 主引用文献 | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 2024
|
|
