6OX1
 
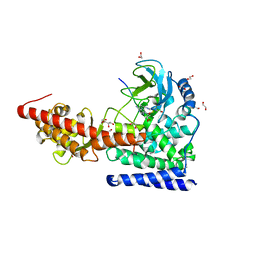 | | SETD3 in Complex with an Actin Peptide with Target Histidine Partially Methylated | | 分子名称: | 1,2-ETHANEDIOL, Actin, cytoplasmic 1, ... | | 著者 | Horton, J.R, Dai, S, Cheng, X. | | 登録日 | 2019-05-13 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.949 Å) | | 主引用文献 | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
6OX5
 
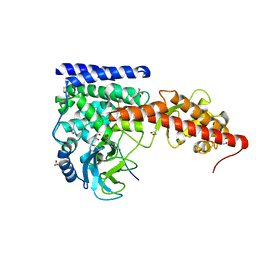 | | A SETD3 Mutant (N255A) in Complex with an Actin Peptide with His73 Replaced with Lysine | | 分子名称: | 1,2-ETHANEDIOL, Actin Peptide, Actin-histidine N-methyltransferase, ... | | 著者 | Horton, J.R, Dai, S, Cheng, X. | | 登録日 | 2019-05-13 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.098 Å) | | 主引用文献 | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
6OX2
 
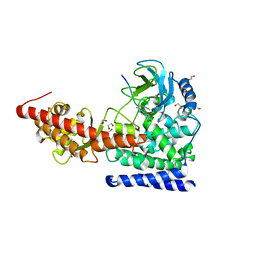 | | SETD3in Complex with an Actin Peptide with the Target Histidine Fully Methylated | | 分子名称: | 1,2-ETHANEDIOL, Actin Peptide, GLYCEROL, ... | | 著者 | Horton, J.R, Dai, S, Cheng, X. | | 登録日 | 2019-05-13 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.089 Å) | | 主引用文献 | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
7RNV
 
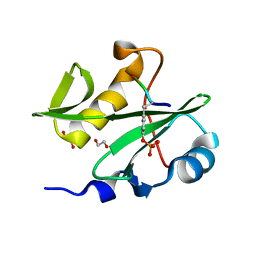 | |
7RNS
 
 | |
7RNU
 
 | |
6OX0
 
 | | SETD3 in Complex with an Actin Peptide with Sinefungin Replacing SAH as Cofactor | | 分子名称: | 1,2-ETHANEDIOL, Actin Peptide, Histone-lysine N-methyltransferase setd3, ... | | 著者 | Horton, J.R, Dai, S, Cheng, X. | | 登録日 | 2019-05-13 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.755 Å) | | 主引用文献 | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
3RGV
 
 | | A single TCR bound to MHCI and MHC II reveals switchable TCR conformers | | 分子名称: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-B alpha chain, ... | | 著者 | Dai, S, Huseby, E, Scott-Browne, J, Rubtsova, K, Pinilla, C, Crawford, F, Marrack, P, Yin, L, Kappler, J.W. | | 登録日 | 2011-04-09 | | 公開日 | 2011-07-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | A Single T Cell Receptor Bound to Major Histocompatibility Complex Class I and Class II Glycoproteins Reveals Switchable TCR Conformers.
Immunity, 35, 2011
|
|
2OG1
 
 | | Crystal Structure of BphD, a C-C hydrolase from Burkholderia xenovorans LB400 | | 分子名称: | 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase, ETHANOL, GLYCEROL, ... | | 著者 | Dai, S, Ke, J, Bolin, J.T. | | 登録日 | 2007-01-04 | | 公開日 | 2007-01-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Kinetic and structural insight into the mechanism of BphD, a C-C bond hydrolase from the biphenyl degradation pathway
Biochemistry, 45, 2006
|
|
8JGU
 
 | |
8JGR
 
 | |
8JGW
 
 | |
8JGX
 
 | |
8JGT
 
 | |
8JGP
 
 | |
8JGO
 
 | |
8JGQ
 
 | |
6DFW
 
 | | TCR 8F10 in complex with IAg7-p8G9E | | 分子名称: | 8F10 alpha chain, 8F10 beta chain, H-2 class II histocompatibility antigen, ... | | 著者 | Wang, Y, Dai, S. | | 登録日 | 2018-05-15 | | 公開日 | 2019-04-17 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | How C-terminal additions to insulin B-chain fragments create superagonists for T cells in mouse and human type 1 diabetes.
Sci Immunol, 4, 2019
|
|
6DFS
 
 | | mouse TCR I.29 in complex with IAg7-p8E9E6ss | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H-2 class II histocompatibility antigen, A-D alpha chain, ... | | 著者 | Wang, Y, Dai, S. | | 登録日 | 2018-05-15 | | 公開日 | 2019-04-17 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | How C-terminal additions to insulin B-chain fragments create superagonists for T cells in mouse and human type 1 diabetes.
Sci Immunol, 4, 2019
|
|
8WA8
 
 | | Human transketolase in complex with phosphite | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, PHOSPHITE ION, ... | | 著者 | Liu, Z, Tittmann, K, Dai, S. | | 登録日 | 2023-09-07 | | 公開日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Multifaceted Role of the Substrate Phosphate Group in Transketolase Catalysis
Acs Catalysis, 14, 2024
|
|
8WA7
 
 | | E.coli transketolase soaked with donor ketose D-fructose | | 分子名称: | 1,2-ETHANEDIOL, 2-[3-[(4-AMINO-2-METHYL-5-PYRIMIDINYL)METHYL]-2-(1,2-DIHYDROXYETHYL)-4-METHYL-1,3-THIAZOL-3-IUM-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, GLYCEROL, ... | | 著者 | Liu, Z, Dai, S, Tittmann, K. | | 登録日 | 2023-09-07 | | 公開日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Multifaceted Role of the Substrate Phosphate Group in Transketolase Catalysis
Acs Catalysis, 14, 2024
|
|
8WA9
 
 | | Human transketolase soaked with donor ketose D-fructose | | 分子名称: | 1,2-ETHANEDIOL, 2-[3-[(4-AMINO-2-METHYL-5-PYRIMIDINYL)METHYL]-2-(1,2-DIHYDROXYETHYL)-4-METHYL-1,3-THIAZOL-3-IUM-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, CALCIUM ION, ... | | 著者 | Liu, Z, Tittmann, K, Dai, S. | | 登録日 | 2023-09-07 | | 公開日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Multifaceted Role of the Substrate Phosphate Group in Transketolase Catalysis
Acs Catalysis, 14, 2024
|
|
8WAA
 
 | | Human transketolase soaked with donor ketose D-xylulose | | 分子名称: | 1,2-ETHANEDIOL, 2-[3-[(4-AMINO-2-METHYL-5-PYRIMIDINYL)METHYL]-2-(1,2-DIHYDROXYETHYL)-4-METHYL-1,3-THIAZOL-3-IUM-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, CALCIUM ION, ... | | 著者 | Liu, Z, Dai, S, Tittmann, K. | | 登録日 | 2023-09-07 | | 公開日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Multifaceted Role of the Substrate Phosphate Group in Transketolase Catalysis
Acs Catalysis, 14, 2024
|
|
8XR3
 
 | |
8XR4
 
 | | Crystal structure of AKRtyl-NADP(H) complex | | 分子名称: | Aldo/keto reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Lin, S, Dai, S, Xiao, Z. | | 登録日 | 2024-01-06 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
