4BBJ
 
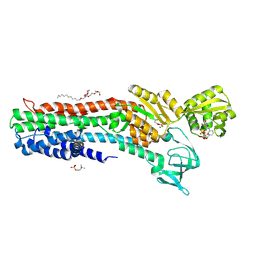 | | Copper-transporting PIB-ATPase in complex with beryllium fluoride representing the E2P state | | 分子名称: | COPPER EFFLUX ATPASE, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Mattle, D, Gourdon, P, Nissen, P. | | 登録日 | 2012-09-25 | | 公開日 | 2013-12-11 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Copper-Transporting P-Type Atpases Use a Unique Ion-Release Pathway
Nat.Struct.Mol.Biol., 21, 2014
|
|
1MXR
 
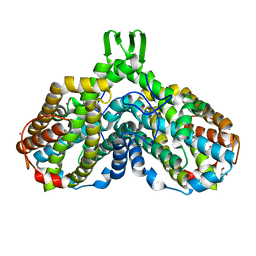 | | High resolution structure of Ribonucleotide reductase R2 from E. coli in its oxidised (Met) form | | 分子名称: | FE (III) ION, GLYCEROL, MERCURY (II) ION, ... | | 著者 | Andersson, M.A, Hogbom, M, Nordlund, P. | | 登録日 | 2002-10-03 | | 公開日 | 2003-03-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Displacement of the tyrosyl radical cofactor in ribonucleotide reductase obtained by single-crystal high-field EPR and 1.4-A x-ray data.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
8EHR
 
 | |
8EHT
 
 | |
8EHS
 
 | |
7ZL4
 
 | | Cryo-EM structure of archaic chaperone-usher Csu pilus of Acinetobacter baumannii | | 分子名称: | CsuA/B | | 著者 | Pakharukova, N, Malmi, H, Tuittila, M, Paavilainen, S, Ghosal, D, Chang, Y.W, Jensen, G.J, Zavialov, A.V. | | 登録日 | 2022-04-13 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.45 Å) | | 主引用文献 | Archaic chaperone-usher pili self-secrete into superelastic zigzag springs.
Nature, 609, 2022
|
|
6C53
 
 | | Cryo-EM structure of the Type 1 pilus rod | | 分子名称: | Type-1 fimbrial protein, A chain | | 著者 | Zheng, W, Wang, F, Luna-Rico, A, Francetic, O, Hultgren, S.J, Egelman, E.H. | | 登録日 | 2018-01-13 | | 公開日 | 2018-01-31 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Functional role of the type 1 pilus rod structure in mediating host-pathogen interactions.
Elife, 7, 2018
|
|
2LTH
 
 | | NMR structure of major ampullate spidroin 1 N-terminal domain at pH 5.5 | | 分子名称: | Major ampullate spidroin 1 | | 著者 | Otikovs, M, Jaudzems, K, Nordling, K, Landreh, M, Rising, A, Askarieh, G, Knight, S, Johansson, J. | | 登録日 | 2012-05-25 | | 公開日 | 2013-11-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Sequential pH-driven dimerization and stabilization of the N-terminal domain enables rapid spider silk formation.
Nat Commun, 5, 2014
|
|
4HQJ
 
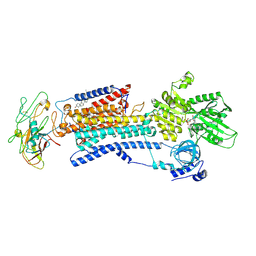 | | Crystal structure of Na+,K+-ATPase in the Na+-bound state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CHOLESTEROL, MAGNESIUM ION, ... | | 著者 | Nyblom, M, Reinhard, L, Gourdon, P, Nissen, P. | | 登録日 | 2012-10-25 | | 公開日 | 2013-10-02 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (4.3 Å) | | 主引用文献 | Crystal structure of Na+, K(+)-ATPase in the Na(+)-bound state.
Science, 342, 2013
|
|
8Q74
 
 | |
5LID
 
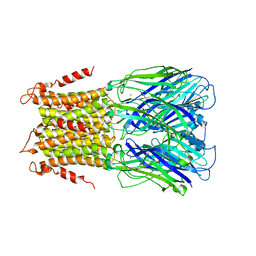 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with bromopromazine | | 分子名称: | Cys-loop ligand-gated ion channel, bromopromazine | | 著者 | Nys, M, Wijckmans, E, Farinha, A, Brams, M, Spurny, R, Ulens, C. | | 登録日 | 2016-07-14 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (4.5 Å) | | 主引用文献 | Allosteric binding site in a Cys-loop receptor ligand-binding domain unveiled in the crystal structure of ELIC in complex with chlorpromazine.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8Q75
 
 | |
8Q76
 
 | |
8Q73
 
 | |
5LG3
 
 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with chlorpromazine | | 分子名称: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, Gamma-aminobutyric-acid receptor subunit beta-1 | | 著者 | Nys, M, Wijckmans, E, Farinha, A, Brams, M, Spurny, R, Ulens, C. | | 登録日 | 2016-07-05 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.567 Å) | | 主引用文献 | Allosteric binding site in a Cys-loop receptor ligand-binding domain unveiled in the crystal structure of ELIC in complex with chlorpromazine.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7PGE
 
 | | copper transporter PcoB | | 分子名称: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Copper resistance protein B, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | 著者 | Li, P, Gourdon, P.E. | | 登録日 | 2021-08-13 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | PcoB is a defense outer membrane protein that facilitates cellular uptake of copper.
Protein Sci., 31, 2022
|
|
7QBZ
 
 | | Crystal structure Cadmium translocating P-type ATPase | | 分子名称: | Cadmium translocating P-type ATPase, MAGNESIUM ION, TETRAFLUOROALUMINATE ION | | 著者 | Groenberg, C, Hu, Q, Wang, K, Gourdon, P. | | 登録日 | 2021-11-21 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Structure and ion-release mechanism of P IB-4 -type ATPases.
Elife, 10, 2021
|
|
7QC0
 
 | | Crystal structure of Cadmium translocating P-type ATPase | | 分子名称: | BERYLLIUM TRIFLUORIDE ION, Cadmium translocating P-type ATPase, MAGNESIUM ION | | 著者 | Groenberg, C, Hu, Q, Wang, K, Gourdon, P. | | 登録日 | 2021-11-21 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | Structure and ion-release mechanism of P IB-4 -type ATPases.
Elife, 10, 2021
|
|
7R0H
 
 | | STRUCTURAL BASIS OF ION UPTAKE IN COPPER-TRANSPORTING P1B-TYPE ATPASES | | 分子名称: | COPPER (II) ION, Putative copper-exporting P-type ATPase A | | 著者 | Salustros, N, Groenberg, C, Wang, K, Gourdon, P. | | 登録日 | 2022-02-02 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.31 Å) | | 主引用文献 | Structural basis of ion uptake in copper-transporting P 1B -type ATPases.
Nat Commun, 13, 2022
|
|
7R0I
 
 | | STRUCTURAL BASIS OF ION UPTAKE IN COPPER-TRANSPORTING P1B-TYPE ATPASES | | 分子名称: | MAGNESIUM ION, POTASSIUM ION, Putative copper-exporting P-type ATPase A | | 著者 | Salustros, N, Groenberg, C, Wang, K, Gourdon, P. | | 登録日 | 2022-02-02 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis of ion uptake in copper-transporting P 1B -type ATPases.
Nat Commun, 13, 2022
|
|
7R0G
 
 | |
6QVD
 
 | |
6QVC
 
 | |
6QV6
 
 | |
6QVB
 
 | |
