2MLO
 
 | | Human CCR2 Membrane-Proximal C-Terminal Region (PRO-C) in a Membrane bound form | | 分子名称: | MCP-1 receptor | | 著者 | Esaki, K, Yoshinaga, S, Tsuji, T, Toda, E, Terashima, Y, Saitoh, T, Kohda, D, Kohno, T, Osawa, M, Ueda, T, Shimada, I, Matsushima, K, Terasawa, H. | | 登録日 | 2014-03-04 | | 公開日 | 2014-10-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for the binding of the membrane-proximal C-terminal region of chemokine receptor CCR2 with the cytosolic regulator FROUNT.
Febs J., 281, 2014
|
|
2MLQ
 
 | | Human CCR2 Membrane-Proximal C-Terminal Region (PRO-C) in a frount bound form | | 分子名称: | MCP-1 receptor | | 著者 | Esaki, K, Yoshinaga, S, Tsuji, T, Toda, E, Terashima, Y, Saitoh, T, Kohda, D, Kohno, T, Osawa, M, Ueda, T, Shimada, I, Matsushima, K, Terasawa, H. | | 登録日 | 2014-03-04 | | 公開日 | 2014-10-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for the binding of the membrane-proximal C-terminal region of chemokine receptor CCR2 with the cytosolic regulator FROUNT.
Febs J., 281, 2014
|
|
2MW8
 
 | |
3WT2
 
 | | Crystal structure of the b'-a' domain of thermophilic fungal protein disulfide isomerase (oxidized form) | | 分子名称: | Protein disulfide-isomerase | | 著者 | Inagaki, K, Satoh, T, Itoh, S.G, Okumura, H, Kato, K. | | 登録日 | 2014-04-02 | | 公開日 | 2014-11-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Redox-dependent conformational transition of catalytic domain of protein disulfide isomerase indicated by crystal structure-based molecular dynamics simulation
Chem.Phys.Lett., 618, 2015
|
|
5XZK
 
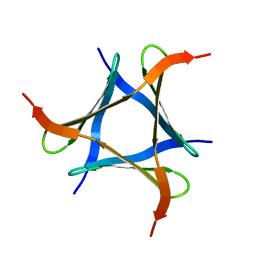 | | Pholiota squarrosa lectin trimer | | 分子名称: | lectin (PhoSL) | | 著者 | Yamasaki, K. | | 登録日 | 2017-07-12 | | 公開日 | 2018-06-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The trimeric solution structure and fucose-binding mechanism of the core fucosylation-specific lectin PhoSL.
Sci Rep, 8, 2018
|
|
4JP8
 
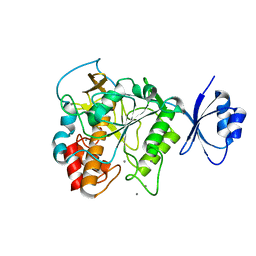 | | Crystal structure of Pro-F17H/S324A | | 分子名称: | CALCIUM ION, Tk-subtilisin | | 著者 | Yuzaki, K, You, D.J, Uehara, R, Koga, Y, Kanaya, S. | | 登録日 | 2013-03-19 | | 公開日 | 2014-01-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Increase in activation rate of Pro-Tk-subtilisin by a single nonpolar-to-polar amino acid substitution at the hydrophobic core of the propeptide domain
Protein Sci., 22, 2013
|
|
2RRK
 
 | |
1GCF
 
 | | NMR STRUCTURE OF THE C-TERMINAL DOMAIN OF THE LIGAND-BINDING REGION OF MURINE GRANULOCYTE COLONY-STIMULATING FACTOR RECEPTOR, 12 STRUCTURES | | 分子名称: | GRANULOCYTE COLONY-STIMULATING FACTOR RECEPTOR | | 著者 | Yamasaki, K, Naito, S, Anaguchi, H, Ohkubo, T, Ota, Y. | | 登録日 | 1997-04-10 | | 公開日 | 1997-10-22 | | 最終更新日 | 2018-03-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of an extracellular domain containing the WSxWS motif of the granulocyte colony-stimulating factor receptor and its interaction with ligand.
Nat.Struct.Biol., 4, 1997
|
|
2KXE
 
 | |
2DWN
 
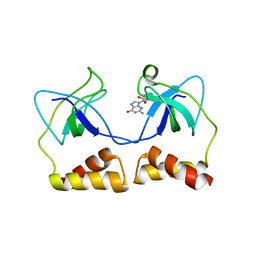 | | Crystal structure of the PriA protein complexed with oligonucleotides | | 分子名称: | DNA (5'-D(*A*G)-3'), Primosomal protein N' | | 著者 | Sasaki, K, Ose, T, Tanaka, T, Masai, H, Maenaka, K, Kohda, D. | | 登録日 | 2006-08-15 | | 公開日 | 2006-11-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2DWM
 
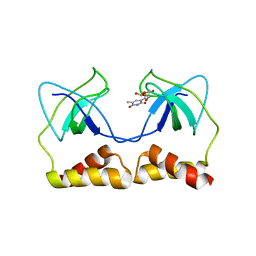 | | Crystal structure of the PriA protein complexed with oligonucleotides | | 分子名称: | 5'-D(*AP*T)-3', Primosomal protein N | | 著者 | Sasaki, K, Ose, T, Tanaka, T, Masai, H, Maenaka, K, Kohda, D. | | 登録日 | 2006-08-15 | | 公開日 | 2006-11-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2DWL
 
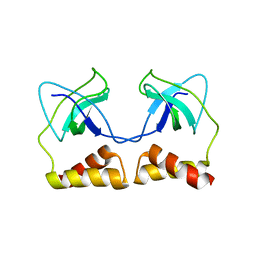 | | Crystal structure of the PriA protein complexed with oligonucleotides | | 分子名称: | 5'-D(*AP*(DC))-3', Primosomal protein N | | 著者 | Sasaki, K, Ose, T, Tanaka, T, Masai, H, Maenaka, K, Kohda, D. | | 登録日 | 2006-08-15 | | 公開日 | 2006-11-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
3WT1
 
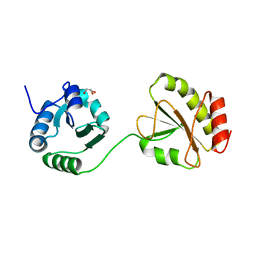 | | Crystal structure of the b'-a' domain of thermophilic fungal protein disulfide isomerase (reduced form) | | 分子名称: | GLYCEROL, Protein disulfide-isomerase | | 著者 | Inagaki, K, Satoh, T, Itoh, S.G, Okumura, H, Kato, K. | | 登録日 | 2014-04-02 | | 公開日 | 2014-11-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Redox-dependent conformational transition of catalytic domain of protein disulfide isomerase indicated by crystal structure-based molecular dynamics simulation
Chem.Phys.Lett., 618, 2015
|
|
3WVF
 
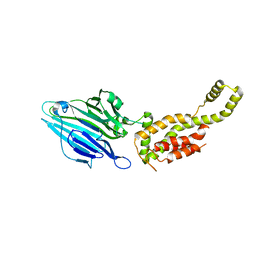 | | Crystal structure of YidC from Escherichia coli | | 分子名称: | Membrane protein insertase YidC | | 著者 | Kumazaki, K, Tsukazaki, T, Kishimoto, T, Ishitani, R, Nureki, O. | | 登録日 | 2014-05-20 | | 公開日 | 2014-12-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Crystal structure of Escherichia coli YidC, a membrane protein chaperone and insertase
Sci Rep, 4, 2014
|
|
1ISP
 
 | | Crystal structure of Bacillus subtilis lipase at 1.3A resolution | | 分子名称: | GLYCEROL, lipase | | 著者 | Kawasaki, K, Kondo, H, Suzuki, M, Ohgiya, S, Tsuda, S. | | 登録日 | 2001-12-19 | | 公開日 | 2002-12-19 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Alternate conformations observed in catalytic serine of Bacillus subtilis lipase determined at 1.3 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2CZQ
 
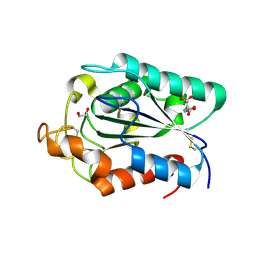 | | A novel cutinase-like protein from Cryptococcus sp. | | 分子名称: | 1,2-ETHANEDIOL, CITRIC ACID, cutinase-like protein | | 著者 | Masaki, K, Kamini, N.R, Ikeda, H, Iefuji, H, Kondo, H, Suzuki, M, Tsuda, S. | | 登録日 | 2005-07-14 | | 公開日 | 2006-07-14 | | 最終更新日 | 2012-06-13 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Crystal structure and enhanced activity of a cutinase-like enzyme from Cryptococcus sp. strain S-2
Proteins, 77, 2009
|
|
3W3D
 
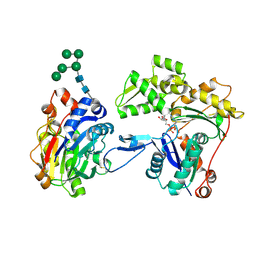 | | Crystal structure of smooth muscle G actin DNase I complex | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Actin, gamma-enteric smooth muscle, ... | | 著者 | Sakabe, N, Sakabe, K, Sasaki, K, Kondo, H, Shimomur, M. | | 登録日 | 2012-12-20 | | 公開日 | 2013-01-30 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Refined structure and solvent network of chicken gizzard G-actin DNase 1 complex at 1.8A resolution
Acta Crystallogr.,Sect.A, 49, 1993
|
|
7WKZ
 
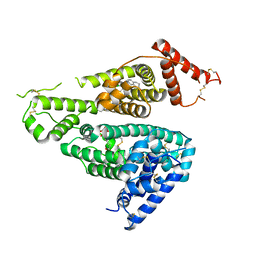 | |
2CWR
 
 | |
1C9F
 
 | |
6LFF
 
 | | transcription factor SATB1 CUTr1 domain in complex with a phosphorothioate DNA | | 分子名称: | DNA (5'-D(*GP*(C7R)P*(PST)P*AP*AP*TP*AP*TP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*(AS)P*(PST)P*(AS)P*(PST)P*TP*AP*GP*C)-3'), DNA-binding protein SATB1 | | 著者 | Akutsu, Y, Kubota, T, Yamasaki, T, Yamasaki, K. | | 登録日 | 2019-12-02 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Enhanced affinity of racemic phosphorothioate DNA with transcription factor SATB1 arising from diastereomer-specific hydrogen bonds and hydrophobic contacts.
Nucleic Acids Res., 48, 2020
|
|
7D6J
 
 | | Human serum albumin complexed with benzbromarone | | 分子名称: | Serum albumin, [3,5-bis(bromanyl)-4-oxidanyl-phenyl]-(2-ethyl-1-benzofuran-3-yl)methanone | | 著者 | Kawai, A, Yamasaki, K. | | 登録日 | 2020-09-30 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.29 Å) | | 主引用文献 | Interaction of Benzbromarone with Subdomains IIIA and IB/IIA on Human Serum Albumin as the Primary and Secondary Binding Regions.
Mol Pharm., 18, 2021
|
|
2RTS
 
 | | Chitin binding domain1 | | 分子名称: | chitinase | | 著者 | Uegaki, K. | | 登録日 | 2013-08-19 | | 公開日 | 2014-04-23 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the chitin-binding domain 1 (ChBD1) of a hyperthermophilic chitinase from Pyrococcus furiosus.
J.Biochem., 155, 2014
|
|
1YSE
 
 | |
1GCC
 
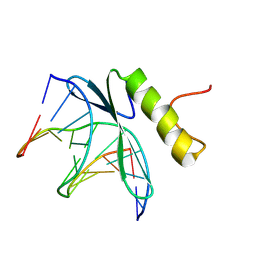 | | SOLUTION NMR STRUCTURE OF THE COMPLEX OF GCC-BOX BINDING DOMAIN OF ATERF1 AND GCC-BOX DNA, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | DNA (5'-D(*GP*CP*TP*GP*GP*CP*GP*GP*CP*TP*A)-3'), DNA (5'-D(*TP*AP*GP*CP*CP*GP*CP*CP*AP*GP*C)-3'), ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 | | 著者 | Yamasaki, K, Allen, M.D, Ohme-Takagi, M, Tateno, M, Suzuki, M. | | 登録日 | 1998-03-13 | | 公開日 | 1999-03-23 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA.
EMBO J., 17, 1998
|
|
