2ISI
 
 | |
4E1J
 
 | | Crystal structure of glycerol kinase in complex with glycerol from Sinorhizobium meliloti 1021 | | 分子名称: | CHLORIDE ION, GLYCEROL, Glycerol kinase, ... | | 著者 | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Siedel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-03-06 | | 公開日 | 2012-03-21 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Crystal structure of glycerol kinase in complex with glycerol from Sinorhizobium meliloti 1021
To be Published
|
|
4DYV
 
 | | Crystal structure of a short-chain dehydrogenase/reductase SDR from Xanthobacter autotrophicus Py2 | | 分子名称: | CHLORIDE ION, Short-chain dehydrogenase/reductase SDR | | 著者 | Agarwal, R, Chamala, S, Evans, B, Foti, R, Hillerich, B, Kar, A, Lafleur, J, Siedel, R, Villigas, G, Zencheck, W, Gizzi, A, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-02-29 | | 公開日 | 2012-03-14 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a short-chain dehydrogenase/reductase SDR from Xanthobacter autotrophicus Py2
To be Published
|
|
4DQX
 
 | | Crystal structure of a short chain dehydrogenase from Rhizobium etli CFN 42 | | 分子名称: | Probable oxidoreductase protein | | 著者 | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Siedel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-02-16 | | 公開日 | 2012-02-29 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of a short chain dehydrogenase from Rhizobium etli CFN 42
To be Published
|
|
4DLL
 
 | | Crystal structure of a 2-hydroxy-3-oxopropionate reductase from Polaromonas sp. JS666 | | 分子名称: | 2-hydroxy-3-oxopropionate reductase, SULFATE ION | | 著者 | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-02-06 | | 公開日 | 2012-02-15 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Crystal structure of a 2-hydroxy-3-oxopropionate reductase from Polaromonas sp. JS666
To be Published
|
|
4DVJ
 
 | | Crystal structure of a putative zinc-dependent alcohol dehydrogenase protein from Rhizobium etli CFN 42 | | 分子名称: | Putative zinc-dependent alcohol dehydrogenase protein | | 著者 | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hellerich, B, Kar, A, Lafleur, J, Siedel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-02-23 | | 公開日 | 2012-03-07 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Crystal structure of a putative zinc-dependent alcohol dehydrogenase protein from Rhizobium etli CFN 42
To be Published
|
|
4DU5
 
 | | Crystal structure of PfkB protein from Polaromonas sp. JS666 | | 分子名称: | CHLORIDE ION, PfkB | | 著者 | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hellerich, B, Kar, A, Lafleur, J, Siedel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-02-21 | | 公開日 | 2012-03-21 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of PfkB protein from Polaromonas sp. JS666
To be Published
|
|
4F3S
 
 | | Crystal structure of periplasmic D-alanine ABC transporter from Salmonella enterica | | 分子名称: | D-ALANINE, GLYCINE, PHOSPHATE ION, ... | | 著者 | Agarwal, R, Chamala, S, Evans, B, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Siedel, R, Villigas, G, Zencheck, W, Foti, R, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-05-09 | | 公開日 | 2012-05-23 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Crystal structure of periplasmic D-alanine ABC transporter from Salmonella enterica
To be Published
|
|
1RTT
 
 | | Crystal structure determination of a putative NADH-dependent reductase using sulfur anomalous signal | | 分子名称: | SULFATE ION, conserved hypothetical protein | | 著者 | Agarwal, R, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2003-12-10 | | 公開日 | 2004-08-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | Structure determination of an FMN reductase from Pseudomonas aeruginosa PA01 using sulfur anomalous signal.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
1SG9
 
 | | Crystal structure of Thermotoga maritima protein HEMK, an N5-glutamine methyltransferase | | 分子名称: | GLUTAMINE, S-ADENOSYLMETHIONINE, hemK protein | | 著者 | Agarwal, R, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2004-02-23 | | 公開日 | 2004-08-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A novel mode of dimerization via formation of a glutamate anhydride crosslink in a protein crystal structure.
Proteins, 71, 2008
|
|
1T3C
 
 | | Clostridium botulinum type E catalytic domain E212Q mutant | | 分子名称: | CHLORIDE ION, ZINC ION, neurotoxin type E | | 著者 | Agarwal, R, Eswaramoorthy, S, Kumaran, D, Binz, T, Swaminathan, S. | | 登録日 | 2004-04-26 | | 公開日 | 2004-06-29 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural analysis of botulinum neurotoxin type E catalytic domain and its mutant Glu212-->Gln reveals the pivotal role of the Glu212 carboxylate in the catalytic pathway
Biochemistry, 43, 2004
|
|
1T3A
 
 | | Crystal structure of Clostridium botulinum neurotoxin type E catalytic domain | | 分子名称: | CHLORIDE ION, ZINC ION, neurotoxin type E | | 著者 | Agarwal, R, Eswaramoorthy, S, Kumaran, D, Binz, T, Swaminathan, S. | | 登録日 | 2004-04-26 | | 公開日 | 2004-06-29 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Structural analysis of botulinum neurotoxin type E catalytic domain and its mutant Glu212-->Gln reveals the pivotal role of the Glu212 carboxylate in the catalytic pathway
Biochemistry, 43, 2004
|
|
1Z2L
 
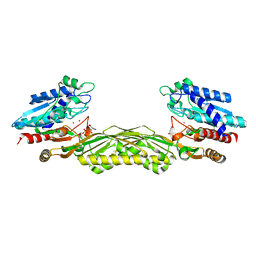 | | Crystal structure of Allantoate-amidohydrolase from E.coli K12 in complex with substrate Allantoate | | 分子名称: | ALLANTOATE ION, Allantoate amidohydrolase, SULFATE ION, ... | | 著者 | Agarwal, R, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2005-03-08 | | 公開日 | 2005-03-22 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural analysis of a ternary complex of allantoate amidohydrolase from Escherichia coli reveals its mechanics.
J.Mol.Biol., 368, 2007
|
|
2A97
 
 | |
1ZL6
 
 | | Crystal structure of Tyr350Ala mutant of Clostridium botulinum neurotoxin E catalytic domain | | 分子名称: | SULFATE ION, ZINC ION, botulinum neurotoxin type E | | 著者 | Agarwal, R, Binz, T, Swaminathan, S. | | 登録日 | 2005-05-05 | | 公開日 | 2005-06-28 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
1ZL5
 
 | | Crystal structure of Glu335Gln mutant of Clostridium botulinum neurotoxin E catalytic domain | | 分子名称: | CHLORIDE ION, botulinum neurotoxin type E | | 著者 | Agarwal, R, Binz, T, Swaminathan, S. | | 登録日 | 2005-05-05 | | 公開日 | 2005-07-05 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
1ZKW
 
 | | Crystal structure of Arg347Ala mutant of botulinum neurotoxin E catalytic domain | | 分子名称: | CHLORIDE ION, ZINC ION, botulinum neurotoxin type E | | 著者 | Agarwal, R, Binz, T, Swaminathan, S. | | 登録日 | 2005-05-04 | | 公開日 | 2005-06-28 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
1ZKX
 
 | | Crystal structure of Glu158Ala/Thr159Ala/Asn160Ala- a triple mutant of Clostridium botulinum neurotoxin E catalytic domain | | 分子名称: | CHLORIDE ION, ZINC ION, botulinum neurotoxin type E | | 著者 | Agarwal, R, Binz, T, Swaminathan, S. | | 登録日 | 2005-05-04 | | 公開日 | 2005-07-05 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
1ZN3
 
 | | Crystal structure of Glu335Ala mutant of Clostridium botulinum neurotoxin type E | | 分子名称: | CHLORIDE ION, ZINC ION, botulinum neurotoxin type E | | 著者 | Agarwal, R, Binz, T, Swaminathan, S. | | 登録日 | 2005-05-11 | | 公開日 | 2005-07-05 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
2A8A
 
 | |
3R64
 
 | |
3R2G
 
 | |
3R31
 
 | |
3R4Q
 
 | |
3R3H
 
 | |
