6LOZ
 
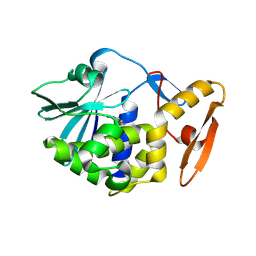 | | crystal structure of alpha-momorcharin in complex with adenine | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, Ribosome-inactivating protein momordin I | | 著者 | Fan, X, Jin, T. | | 登録日 | 2020-01-07 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
6LP0
 
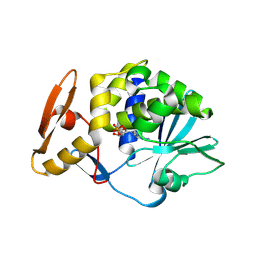 | | crystal structure of alpha-momorcharin in complex with AMP | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE MONOPHOSPHATE, Ribosome-inactivating protein momordin I | | 著者 | Fan, X, Jin, T. | | 登録日 | 2020-01-07 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.519 Å) | | 主引用文献 | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
6LOR
 
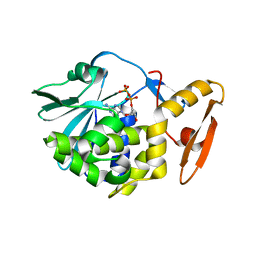 | | crystal structure of alpha-momorcharin in complex with ADP | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, Ribosome-inactivating protein momordin I | | 著者 | Fan, X, Jin, T. | | 登録日 | 2020-01-07 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
6LOW
 
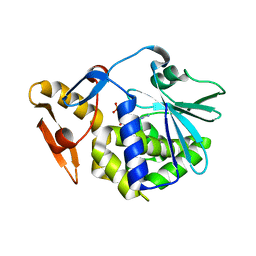 | | crystal structure of alpha-momorcharin in complex with GMP | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GUANOSINE-5'-MONOPHOSPHATE, Ribosome-inactivating protein momordin I | | 著者 | Fan, X, Jin, T. | | 登録日 | 2020-01-07 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
6LOQ
 
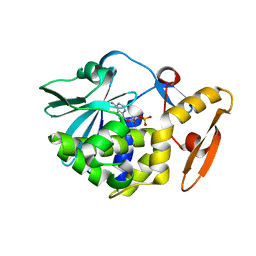 | | crystal structure of alpha-momorcharin in complex with cAMP | | 分子名称: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Ribosome-inactivating protein momordin I | | 著者 | Fan, X, Jin, T. | | 登録日 | 2020-01-07 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.331 Å) | | 主引用文献 | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
6LOY
 
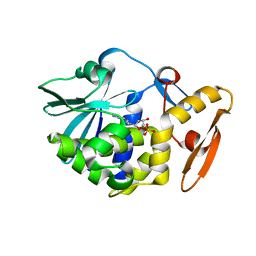 | | crystal structure of alpha-momorcharin in complex with dAMP | | 分子名称: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Ribosome-inactivating protein momordin I | | 著者 | Fan, X, Jin, T. | | 登録日 | 2020-01-07 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
6LOV
 
 | |
5VYK
 
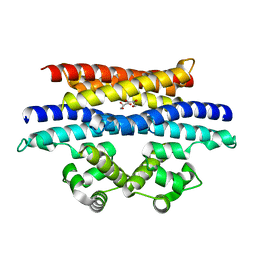 | | Crystal structure of the BRS domain of BRAF in complex with the CC-SAM domain of KSR1 | | 分子名称: | Chimera protein of BRS domain of BRAF and CC-SAM domain of KSR1,Serine/threonine-protein kinase B-raf, GLYCEROL | | 著者 | Maisonneuve, P, Kurinov, I, Marullo, S.A, Lavoie, H, Thevakumaran, N, Sahmi, M, Jin, T, Therrien, M, SIcheri, F. | | 登録日 | 2017-05-25 | | 公開日 | 2018-02-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.749 Å) | | 主引用文献 | MEK drives BRAF activation through allosteric control of KSR proteins.
Nature, 554, 2018
|
|
5VR3
 
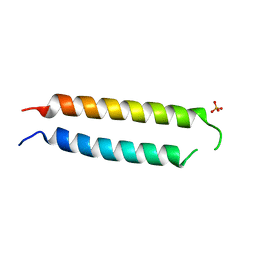 | | Crystal structure of the BRS domain of BRAF | | 分子名称: | BRAF, SULFATE ION | | 著者 | Thevakumaran, N, Maisonneuve, P, Kurinov, I, Lavoie, H, Marullo, S.A, Sahmi, M, Jin, T, Therrien, M, Sicheri, F. | | 登録日 | 2017-05-10 | | 公開日 | 2018-02-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.102 Å) | | 主引用文献 | MEK drives BRAF activation through allosteric control of KSR proteins.
Nature, 554, 2018
|
|
5YEV
 
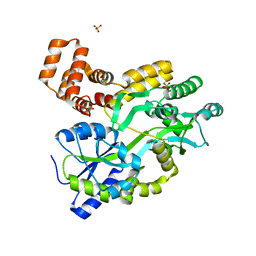 | | Murine DR3 death domain | | 分子名称: | SULFATE ION, TNFRSF25 death domain | | 著者 | Yin, X, Jin, T. | | 登録日 | 2017-09-19 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
5YGS
 
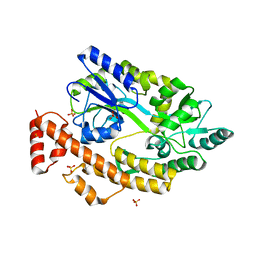 | | Human TNFRSF25 death domain | | 分子名称: | Human TNRSF25 death domain, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Yin, X, Jin, T. | | 登録日 | 2017-09-26 | | 公開日 | 2018-10-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.691 Å) | | 主引用文献 | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
5ZNY
 
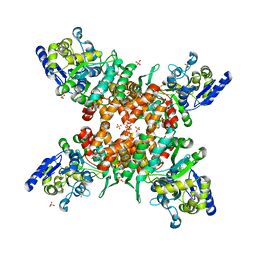 | | Structure of mDR3_DD-C363G with MBP tag | | 分子名称: | Maltose-binding periplasmic protein,Tumor necrosis factor receptor superfamily, member 25, SULFATE ION | | 著者 | Yin, X, Jin, T. | | 登録日 | 2018-04-11 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.74 Å) | | 主引用文献 | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
5ZTC
 
 | |
7FCQ
 
 | |
7FCP
 
 | |
6JOX
 
 | | triosephosphate isomerase-scylla paramamosain | | 分子名称: | Triosephosphate isomerase | | 著者 | Xia, F, Jin, T. | | 登録日 | 2019-03-25 | | 公開日 | 2019-12-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.803 Å) | | 主引用文献 | Crystal Structure Analysis and Conformational Epitope Mutation of Triosephosphate Isomerase, a Mud Crab Allergen.
J.Agric.Food Chem., 67, 2019
|
|
6JYM
 
 | |
7EU2
 
 | |
7F4W
 
 | |
7VZO
 
 | |
7WBO
 
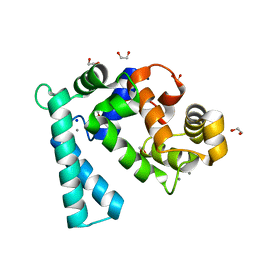 | | Crystal structure of Sarcoplasmic Calcium-Binding Protein from Scylla paramamosain | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, SODIUM ION, ... | | 著者 | Chen, Y, Jin, T, Liu, G. | | 登録日 | 2021-12-17 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structure Analysis of Sarcoplasmic-Calcium-Binding Protein: An Allergen in Scylla paramamosain.
J.Agric.Food Chem., 71, 2023
|
|
2RE9
 
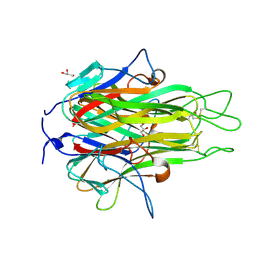 | | Crystal structure of TL1A at 2.1 A | | 分子名称: | GLYCEROL, MAGNESIUM ION, TNF superfamily ligand TL1A | | 著者 | Jin, T.C, Guo, F, Kim, S, Howard, A.J, Zhang, Y.Z. | | 登録日 | 2007-09-25 | | 公開日 | 2007-10-09 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | X-ray crystal structure of TNF ligand family member TL1A at 2.1 A.
Biochem.Biophys.Res.Commun., 364, 2007
|
|
3RLO
 
 | |
3RN2
 
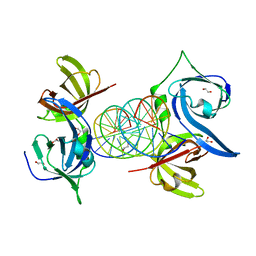 | | Structural Basis of Cytosolic DNA Recognition by Innate Immune Receptors | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*TP*CP*AP*AP*AP*GP*AP*TP*CP*TP*TP*TP*GP*AP*TP*GP*G)-3'), Interferon-inducible protein AIM2 | | 著者 | Jin, T.C, Xiao, T. | | 登録日 | 2011-04-21 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor.
Immunity, 36, 2012
|
|
3RLN
 
 | |
