7VY5
 
 | | Coxsackievirus B3 (VP3-234Q) incubation with CD55 at pH7.4 | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | 著者 | Wang, Q.L, Liu, C.C. | | 登録日 | 2021-11-13 | | 公開日 | 2022-01-19 | | 最終更新日 | 2022-08-10 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VXZ
 
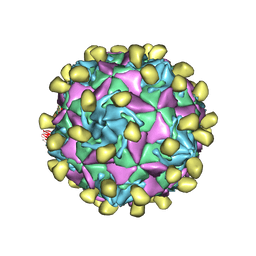 | |
7VYK
 
 | |
7VYL
 
 | |
7VXH
 
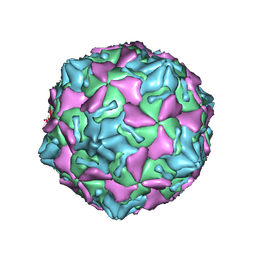 | | Coxsackievirus B3 full particle at pH7.4 (VP3-234Q) | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | 著者 | Wang, Q.L, Liu, C.C. | | 登録日 | 2021-11-12 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VYM
 
 | |
7VY0
 
 | | Coxsackievirus B3 full particle at pH7.4 (VP3-234N) | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | 著者 | Wang, Q.L, Liu, C.C. | | 登録日 | 2021-11-13 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VY6
 
 | | Coxsackievirus B3(VP3-234N) incubate with CD55 at pH7.4 | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | 著者 | Wang, Q.L, Liu, C.C. | | 登録日 | 2021-11-13 | | 公開日 | 2022-01-19 | | 最終更新日 | 2022-08-10 | | 実験手法 | ELECTRON MICROSCOPY (3.02 Å) | | 主引用文献 | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7V61
 
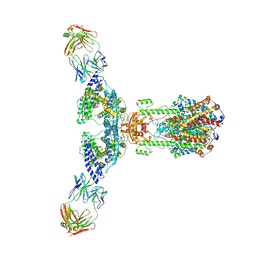 | | ACE2 -Targeting Monoclonal Antibody as Potent and Broad-Spectrum Coronavirus Blocker | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3E8, ... | | 著者 | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | 登録日 | 2021-08-18 | | 公開日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | ACE2-targeting monoclonal antibody as potent and broad-spectrum coronavirus blocker.
Signal Transduct Target Ther, 6, 2021
|
|
1O2F
 
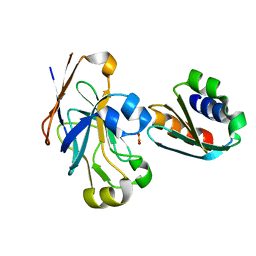 | | COMPLEX OF ENZYME IIAGLC AND IIBGLC PHOSPHOCARRIER PROTEIN HPR FROM ESCHERICHIA COLI NMR, RESTRAINED REGULARIZED MEAN STRUCTURE | | 分子名称: | PHOSPHITE ION, PTS system, glucose-specific IIA component, ... | | 著者 | Clore, G.M, Cai, M, Williams, D.C. | | 登録日 | 2003-03-11 | | 公開日 | 2003-05-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the Phosphoryl Transfer Complex between the Signal-transducing Protein IIAGlucose and the Cytoplasmic Domain of the Glucose Transporter IICBGlucose of the Escherichia coli Glucose Phosphotransferase System.
J.Biol.Chem., 278, 2003
|
|
6BEA
 
 | |
1T05
 
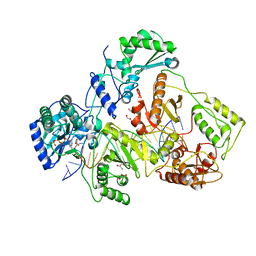 | | HIV-1 reverse transcriptase crosslinked to template-primer with tenofovir-diphosphate bound as the incoming nucleotide substrate | | 分子名称: | GLYCEROL, MAGNESIUM ION, POL polyprotein, ... | | 著者 | Tuske, S, Sarafianos, S.G, Ding, J, Arnold, E. | | 登録日 | 2004-04-07 | | 公開日 | 2004-05-11 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structures of HIV-1 RT-DNA complexes before and after incorporation of the anti-AIDS drug tenofovir
Nat.Struct.Mol.Biol., 11, 2004
|
|
7W14
 
 | |
7W17
 
 | |
4JDL
 
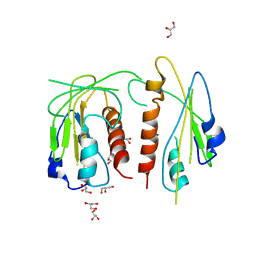 | |
1T03
 
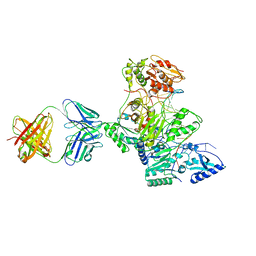 | | HIV-1 reverse transcriptase crosslinked to tenofovir terminated template-primer (complex P) | | 分子名称: | MAGNESIUM ION, POL polyprotein, Synthetic oligonucleotide primer, ... | | 著者 | Tuske, S, Sarafianos, S.G, Ding, J, Arnold, E. | | 登録日 | 2004-04-07 | | 公開日 | 2004-05-11 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure of HIV-1 RT-DNA complexes before and after incorporation of the anti-AIDS drug tenofovir
Nat.Struct.Mol.Biol., 11, 2004
|
|
7BZ7
 
 | | Template lasso peptide C24 mutant F15Y | | 分子名称: | lasso peptide | | 著者 | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | 登録日 | 2020-04-27 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
5DOO
 
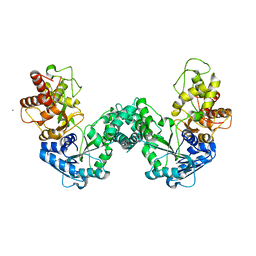 | | The structure of PKMT2 from Rickettsia typhi | | 分子名称: | CALCIUM ION, protein lysine methyltransferase 2 | | 著者 | Noinaj, N, Abeykoon, A, He, Y, Yang, D.C, Buchanan, S.K. | | 登録日 | 2015-09-11 | | 公開日 | 2016-08-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.133 Å) | | 主引用文献 | Structural Insights into Substrate Recognition and Catalysis in Outer Membrane Protein B (OmpB) by Protein-lysine Methyltransferases from Rickettsia.
J.Biol.Chem., 291, 2016
|
|
5DPD
 
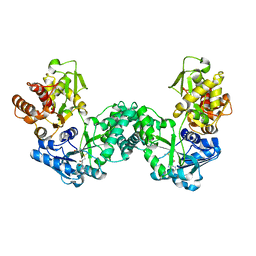 | | The structure of PKMT1 from Rickettsia prowazekii in complex with AdoMet | | 分子名称: | S-ADENOSYLMETHIONINE, protein lysine methyltransferase 1 | | 著者 | Noinaj, N, Abeykoon, A, He, Y, Yang, D.C, Buchanan, S.K. | | 登録日 | 2015-09-12 | | 公開日 | 2016-08-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Insights into Substrate Recognition and Catalysis in Outer Membrane Protein B (OmpB) by Protein-lysine Methyltransferases from Rickettsia.
J.Biol.Chem., 291, 2016
|
|
5DO0
 
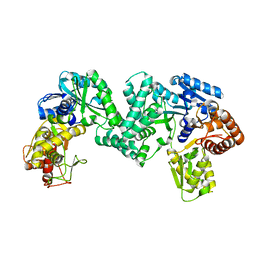 | | The structure of PKMT1 from Rickettsia prowazekii | | 分子名称: | protein lysine methyltransferase 1 | | 著者 | Noinaj, N, Abeykoon, A, He, Y, Yang, D.C, Buchanan, S.K. | | 登録日 | 2015-09-10 | | 公開日 | 2016-08-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural Insights into Substrate Recognition and Catalysis in Outer Membrane Protein B (OmpB) by Protein-lysine Methyltransferases from Rickettsia.
J.Biol.Chem., 291, 2016
|
|
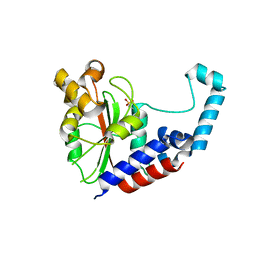
3EIR
 
 | |
3EIT
 
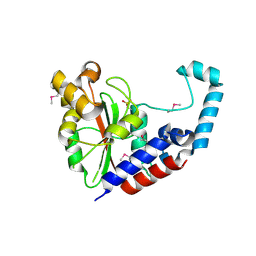 | |
7YP1
 
 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 10E4 (localized refinement) | | 分子名称: | 10E4 heavy chain, 10E4 light chain, EBV gH, ... | | 著者 | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | 登録日 | 2022-08-02 | | 公開日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
7YOY
 
 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAbs 3E8 and 5E3 (localized refinement) | | 分子名称: | 3E8 heavy chain, 3E8 light chain, 5E3 heavy chain, ... | | 著者 | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | 登録日 | 2022-08-02 | | 公開日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3.64 Å) | | 主引用文献 | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
7YP2
 
 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 6H2 (localized refinement) | | 分子名称: | 6H2 heavy chain, 6H2 light chain, Envelope glycoprotein H | | 著者 | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | 登録日 | 2022-08-02 | | 公開日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
