1C56
 
 | |
1CKX
 
 | | Cystic fibrosis transmembrane conductance regulator: Solution structures of peptides based on the Phe508 region, the most common site of disease-causing Delta-F508 mutation | | 分子名称: | Cystic fibrosis transmembrane conductance regulator (CFTR) | | 著者 | Massiah, M.A, Ko, Y.H, Pedersen, P.L, Mildvan, A.S. | | 登録日 | 1999-04-26 | | 公開日 | 1999-05-04 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Cystic fibrosis transmembrane conductance regulator: solution structures of peptides based on the Phe508 region, the most common site of disease-causing DeltaF508 mutation.
Biochemistry, 38, 1999
|
|
1AOS
 
 | |
1CKZ
 
 | | CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR: SOLUTION STRUCTURES OF PEPTIDES BASED ON THE PHE508 REGION, THE MOST COMMON SITE OF DISEASE-CAUSING DELTA-F508 MUTATION | | 分子名称: | PROTEIN (CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR (CFTR)) | | 著者 | Massiah, M.A, Ko, Y.H, Pedersen, P.L, Mildvan, A.S. | | 登録日 | 1999-04-26 | | 公開日 | 1999-05-04 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Cystic fibrosis transmembrane conductance regulator: solution structures of peptides based on the Phe508 region, the most common site of disease-causing DeltaF508 mutation.
Biochemistry, 38, 1999
|
|
1CKY
 
 | | CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR: SOLUTION STRUCTURES OF PEPTIDES BASED ON THE PHE508 REGION, THE MOST COMMON SITE OF DISEASE-CAUSING DELTA-F508 MUTATION | | 分子名称: | PROTEIN (CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR (CFTR)) | | 著者 | Massiah, M.A, Ko, Y.H, Pedersen, P.L, Mildvan, A.S. | | 登録日 | 1999-04-26 | | 公開日 | 1999-05-04 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Cystic fibrosis transmembrane conductance regulator: solution structures of peptides based on the Phe508 region, the most common site of disease-causing DeltaF508 mutation.
Biochemistry, 38, 1999
|
|
1A7A
 
 | | STRUCTURE OF HUMAN PLACENTAL S-ADENOSYLHOMOCYSTEINE HYDROLASE: DETERMINATION OF A 30 SELENIUM ATOM SUBSTRUCTURE FROM DATA AT A SINGLE WAVELENGTH | | 分子名称: | (1'R,2'S)-9-(2-HYDROXY-3'-KETO-CYCLOPENTEN-1-YL)ADENINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, S-ADENOSYLHOMOCYSTEINE HYDROLASE | | 著者 | Turner, M.A, Yuan, C.-S, Borchardt, R.T, Hershfield, M.S, Smith, G.D, Howell, P.L. | | 登録日 | 1998-03-10 | | 公開日 | 1999-04-20 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure determination of selenomethionyl S-adenosylhomocysteine hydrolase using data at a single wavelength.
Nat.Struct.Biol., 5, 1998
|
|
1C55
 
 | |
1DL2
 
 | | CRYSTAL STRUCTURE OF CLASS I ALPHA-1,2-MANNOSIDASE FROM SACCHAROMYCES CEREVISIAE AT 1.54 ANGSTROM RESOLUTION | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CLASS I ALPHA-1,2-MANNOSIDASE, ... | | 著者 | Vallee, F, Lipari, F, Yip, P, Herscovics, A, Howell, P.L. | | 登録日 | 1999-12-08 | | 公開日 | 2000-02-18 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Crystal structure of a class I alpha1,2-mannosidase involved in N-glycan processing and endoplasmic reticulum quality control.
EMBO J., 19, 2000
|
|
1B3C
 
 | | SOLUTION STRUCTURE OF A BETA-NEUROTOXIN FROM THE NEW WORLD SCORPION CENTRUROIDES SCULPTURATUS EWING | | 分子名称: | PROTEIN (NEUROTOXIN CSE-I) | | 著者 | Jablonsky, M.J, Jackson, P.L, Trent, J.O, Watt, D.D, Krishna, N.R. | | 登録日 | 1998-12-08 | | 公開日 | 1998-12-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a beta-neurotoxin from the New World scorpion Centruroides sculpturatus Ewing.
Biochem.Biophys.Res.Commun., 254, 1999
|
|
1EDS
 
 | | SOLUTION STRUCTURE OF INTRADISKAL LOOP 1 OF BOVINE RHODOPSIN (RHODOPSIN RESIDUES 92-123) | | 分子名称: | RHODOPSIN | | 著者 | Yeagle, P.L, Salloum, A, Chopra, A, Bhawsar, N, Ali, L. | | 登録日 | 2000-01-28 | | 公開日 | 2000-08-09 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structures of the intradiskal loops and amino terminus of the G-protein receptor, rhodopsin.
J.Pept.Res., 55, 2000
|
|
1EDW
 
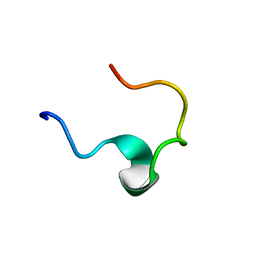 | | SOLUTION STRUCTURE OF THIRD INTRADISKAL LOOP OF BOVINE RHODOPSIN (RESIDUES 268-293) | | 分子名称: | RHODOPSIN | | 著者 | Yeagle, P.L, Salloum, A, Chopra, A, Bhawsar, N, Ali, L. | | 登録日 | 2000-01-28 | | 公開日 | 2000-08-09 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structures of the intradiskal loops and amino terminus of the G-protein receptor, rhodopsin.
J.Pept.Res., 55, 2000
|
|
1EDX
 
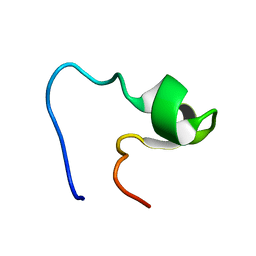 | | SOLUTION STRUCTURE OF AMINO TERMINUS OF BOVINE RHODOPSIN (RESIDUES 1-40) | | 分子名称: | RHODOPSIN | | 著者 | Yeagle, P.L, Salloum, A, Chopra, A, Bhawsar, N, Ali, L. | | 登録日 | 2000-01-28 | | 公開日 | 2000-08-09 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structures of the intradiskal loops and amino terminus of the G-protein receptor, rhodopsin.
J.Pept.Res., 55, 2000
|
|
1EDV
 
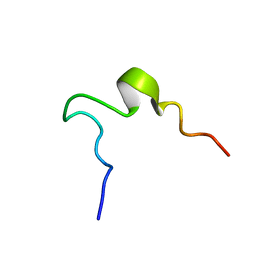 | | SOLUTION STRUCTURE OF 2ND INTRADISKAL LOOP OF BOVINE RHODOPSIN (RESIDUES 172-205) | | 分子名称: | RHODOPSIN | | 著者 | Yeagle, P.L, Salloum, A, Chopra, A, Bhawsar, N, Ali, L. | | 登録日 | 2000-01-28 | | 公開日 | 2000-08-09 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structures of the intradiskal loops and amino terminus of the G-protein receptor, rhodopsin.
J.Pept.Res., 55, 2000
|
|
1CKW
 
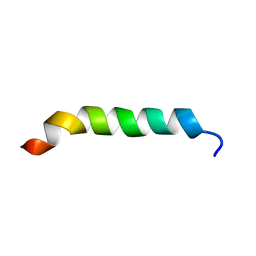 | | CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR: SOLUTION STRUCTURES OF PEPTIDES BASED ON THE PHE508 REGION, THE MOST COMMON SITE OF DISEASE-CAUSING DELTA-F508 MUTATION | | 分子名称: | PROTEIN (CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR (CFTR)) | | 著者 | Massiah, M.A, Ko, Y.H, Pedersen, P.L, Mildvan, A.S. | | 登録日 | 1999-04-26 | | 公開日 | 1999-05-04 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Cystic fibrosis transmembrane conductance regulator: solution structures of peptides based on the Phe508 region, the most common site of disease-causing DeltaF508 mutation.
Biochemistry, 38, 1999
|
|
1EZG
 
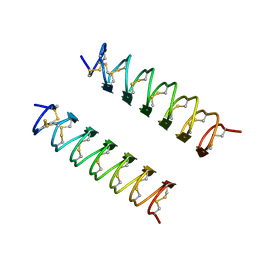 | | CRYSTAL STRUCTURE OF ANTIFREEZE PROTEIN FROM THE BEETLE, TENEBRIO MOLITOR | | 分子名称: | THERMAL HYSTERESIS PROTEIN ISOFORM YL-1 | | 著者 | Liou, Y.-C, Tocilj, A, Davies, P.L, Jia, Z. | | 登録日 | 2000-05-10 | | 公開日 | 2000-08-09 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Mimicry of ice structure by surface hydroxyls and water of a beta-helix antifreeze protein.
Nature, 406, 2000
|
|
1UH5
 
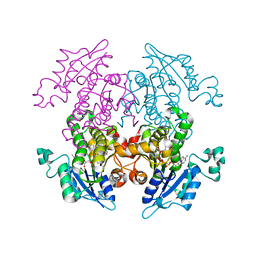 | | Crystal Structure of Enoyl-ACP Reductase with Triclosan at 2.2angstroms | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN, enoyl-ACP reductase | | 著者 | Swarnamukhi, P.L, Kapoor, M, Surolia, N, Surolia, A, Suguna, K. | | 登録日 | 2003-06-24 | | 公開日 | 2004-09-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis for the variation in triclosan affinity to enoyl reductases.
J.Mol.Biol., 343, 2004
|
|
1V35
 
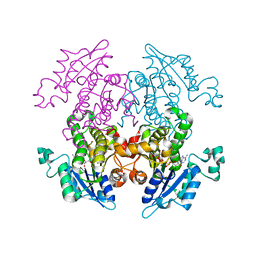 | | Crystal Structure of Eoyl-ACP Reductase with NADH | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, enoyl-ACP reductase | | 著者 | SwarnaMukhi, P.L, Kapoor, M, surolia, N, Surolia, A, Suguna, K. | | 登録日 | 2003-10-28 | | 公開日 | 2004-09-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for the variation in triclosan affinity to enoyl reductases.
J.Mol.Biol., 343, 2004
|
|
4V29
 
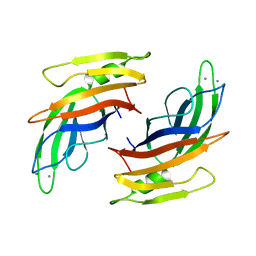 | |
1F5J
 
 | | CRYSTAL STRUCTURE OF XYNB, A HIGHLY THERMOSTABLE BETA-1,4-XYLANASE FROM DICTYOGLOMUS THERMOPHILUM RT46B.1, AT 1.8 A RESOLUTION | | 分子名称: | BETA-1,4-XYLANASE, SULFATE ION | | 著者 | McCarthy, A.A, Baker, E.N. | | 登録日 | 2000-07-26 | | 公開日 | 2000-11-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of XynB, a highly thermostable beta-1,4-xylanase from Dictyoglomus thermophilum Rt46B.1, at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
5XBJ
 
 | |
5J6Y
 
 | |
8IM8
 
 | | Crystal structure of Periplasmic alpha-amylase (MalS) from E.coli | | 分子名称: | CALCIUM ION, Periplasmic alpha-amylase | | 著者 | An, Y, Park, J.T, Park, K.H, Woo, E.J. | | 登録日 | 2023-03-06 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-06-14 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The Distinctive Permutated Domain Structure of Periplasmic alpha-Amylase (MalS) from Glycoside Hydrolase Family 13 Subfamily 19.
Molecules, 28, 2023
|
|
2ZE2
 
 | | Crystal structure of L100I/K103N mutant HIV-1 reverse transcriptase (RT) in complex with TMC278 (rilpivirine), a non-nucleoside RT inhibitor | | 分子名称: | 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | 著者 | Das, K, Bauman, J.D, Clark Jr, A.D, Shatkin, A.J, Arnold, E. | | 登録日 | 2007-12-05 | | 公開日 | 2008-02-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: Strategic flexibility explains potency against resistance mutations.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2ZD1
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (RT) in Complex with TMC278 (Rilpivirine), A Non-nucleoside RT Inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, Reverse transcriptase/ribonuclease H, ... | | 著者 | Das, K, Bauman, J.D, Clark Jr, A.D, Shatkin, A.J, Arnold, E. | | 登録日 | 2007-11-16 | | 公開日 | 2008-02-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: Strategic flexibility explains potency against resistance mutations.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BGR
 
 | | Crystal structure of K103N/Y181C mutant HIV-1 reverse transcriptase (RT) in complex with TMC278 (Rilpivirine), a non-nucleoside RT inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, Reverse transcriptase/ribonuclease H, ... | | 著者 | Das, K, Bauman, J.D, Clark Jr, A.D, Shatkin, A.J, Arnold, E. | | 登録日 | 2007-11-27 | | 公開日 | 2008-02-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: Strategic flexibility explains potency against resistance mutations.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
