5QND
 
 | |
5QLP
 
 | |
5QNT
 
 | |
5QM4
 
 | |
5QO9
 
 | |
5QMK
 
 | |
5QMZ
 
 | |
5QNE
 
 | |
5QNU
 
 | |
5QOA
 
 | |
1O2F
 
 | | COMPLEX OF ENZYME IIAGLC AND IIBGLC PHOSPHOCARRIER PROTEIN HPR FROM ESCHERICHIA COLI NMR, RESTRAINED REGULARIZED MEAN STRUCTURE | | 分子名称: | PHOSPHITE ION, PTS system, glucose-specific IIA component, ... | | 著者 | Clore, G.M, Cai, M, Williams, D.C. | | 登録日 | 2003-03-11 | | 公開日 | 2003-05-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the Phosphoryl Transfer Complex between the Signal-transducing Protein IIAGlucose and the Cytoplasmic Domain of the Glucose Transporter IICBGlucose of the Escherichia coli Glucose Phosphotransferase System.
J.Biol.Chem., 278, 2003
|
|
6KLI
 
 | | Crystal Structure of the Zea Mays laccase 3 complexed with sinapyl | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(1E)-3-hydroxyprop-1-en-1-yl]-2,6-dimethoxyphenol, ... | | 著者 | Xie, T, Liu, Z.C, Wang, G.G. | | 登録日 | 2019-07-30 | | 公開日 | 2020-01-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for monolignol oxidation by a maize laccase.
Nat.Plants, 6, 2020
|
|
6KLJ
 
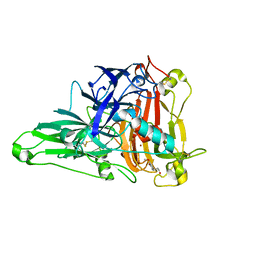 | | Crystal Structure of the Zea Mays laccase 3 complexed with coniferyl | | 分子名称: | (2E)-3-(4-hydroxy-3-methoxyphenyl)prop-2-enal, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Xie, T, Liu, Z.C, Wang, G.G. | | 登録日 | 2019-07-30 | | 公開日 | 2020-01-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.998 Å) | | 主引用文献 | Structural basis for monolignol oxidation by a maize laccase.
Nat.Plants, 6, 2020
|
|
6KLG
 
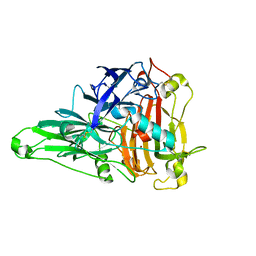 | | Crystal Structure of the Zea Mays laccase 3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | 著者 | Xie, T, Liu, Z.C, Wang, G.G. | | 登録日 | 2019-07-30 | | 公開日 | 2020-01-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for monolignol oxidation by a maize laccase.
Nat.Plants, 6, 2020
|
|
7VUN
 
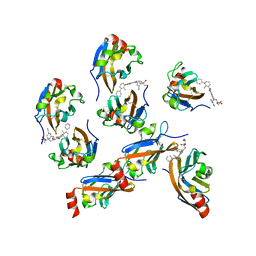 | | Design, modification, evaluation and cocrystal studies of novel phthalimides regulating PD-1/PD-L1 interaction | | 分子名称: | (2~{S},3~{S})-2-[[6-[(3-cyanophenyl)methoxy]-2-(2-methyl-3-phenyl-phenyl)-1,3-bis(oxidanylidene)isoindol-5-yl]methylamino]-3-oxidanyl-butanoic acid, Programmed cell death 1 ligand 1 | | 著者 | Cheng, Y, Sun, C.L, Chen, M.R, Yang, P, Xiao, Y.B. | | 登録日 | 2021-11-03 | | 公開日 | 2022-09-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.701 Å) | | 主引用文献 | Novel phthalimides regulating PD-1/PD-L1 interaction as potential immunotherapy agents.
Acta Pharm Sin B, 12, 2022
|
|
7XC5
 
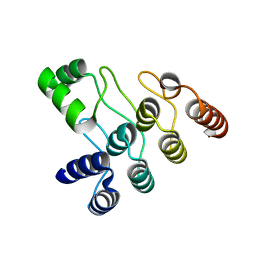 | | Crystal structure of the ANK domain of CLPB | | 分子名称: | Isoform 2 of Caseinolytic peptidase B protein homolog | | 著者 | Liu, Y, Wu, D, Lu, G, Gao, N, Lin, J. | | 登録日 | 2022-03-23 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Comprehensive structural characterization of the human AAA+ disaggregase CLPB in the apo- and substrate-bound states reveals a unique mode of action driven by oligomerization.
Plos Biol., 21, 2023
|
|
3OQU
 
 | | Crystal structure of native abscisic acid receptor PYL9 with ABA | | 分子名称: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL9 | | 著者 | Zhang, X, Zhang, Q, Chen, Z. | | 登録日 | 2010-09-04 | | 公開日 | 2011-09-14 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Structural Insights into the Abscisic Acid Stereospecificity by the ABA Receptors PYR/PYL/RCAR
Plos One, 8, 2013
|
|
7VJS
 
 | | Human AlkB homolog ALKBH6 in complex with Tris and Ni | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 6, NICKEL (II) ION | | 著者 | Ma, L, Chen, Z. | | 登録日 | 2021-09-28 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.792 Å) | | 主引用文献 | Structural insights into the interactions and epigenetic functions of human nucleic acid repair protein ALKBH6.
J.Biol.Chem., 298, 2022
|
|
7VJV
 
 | |
7EP0
 
 | | Crystal structure of ZYG11B bound to GSTE degron | | 分子名称: | Protein zyg-11 homolog B, sodium 3,3'-(1E,1'E)-biphenyl-4,4'-diylbis(diazene-2,1-diyl)bis(4-aminonaphthalene-1-sulfonate) | | 著者 | Yan, X, Li, Y. | | 登録日 | 2021-04-26 | | 公開日 | 2021-07-14 | | 最終更新日 | 2021-09-01 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Molecular basis for recognition of Gly/N-degrons by CRL2 ZYG11B and CRL2 ZER1 .
Mol.Cell, 81, 2021
|
|
7EP3
 
 | |
7EP2
 
 | |
7EP5
 
 | |
7EP1
 
 | |
7EP4
 
 | |
