8VNO
 
 | |
8VMW
 
 | |
8VN7
 
 | |
8VNH
 
 | |
8VNT
 
 | |
7WS6
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | 分子名称: | 510A5 heavy chain, 510A5 light chain, Spike protein S1 | | 著者 | Guo, H, Gao, Y, Ji, X, Yang, H. | | 登録日 | 2022-01-28 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WSA
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1 | | 著者 | Guo, H, Gao, Y, Ji, X, Yang, H. | | 登録日 | 2022-01-28 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS2
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | 分子名称: | 510A5 heavy chain, 510A5 light chain, Spike protein S1 | | 著者 | Guo, H, Gao, Y, Ji, X, Yang, H. | | 登録日 | 2022-01-28 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS8
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Guo, H, Gao, Y, Ji, X, Yang, H. | | 登録日 | 2022-01-28 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS0
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 510A5 heavy chain, ... | | 著者 | Guo, H, Gao, Y, Ji, X, Yang, H. | | 登録日 | 2022-01-28 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS7
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 510A5 heavy chain, 510A5 light chain, ... | | 著者 | Guo, H, Gao, Y, Ji, X, Yang, H. | | 登録日 | 2022-01-28 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS9
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Guo, H, Gao, Y, Ji, X, Yang, H. | | 登録日 | 2022-01-28 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS4
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 510A5 heavy chain, ... | | 著者 | Guo, H, Gao, Y, Ji, X, Yang, H. | | 登録日 | 2022-01-28 | | 公開日 | 2022-06-01 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS3
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 510A5 heavy chain, ... | | 著者 | Guo, H, Gao, Y, Ji, X, Yang, H. | | 登録日 | 2022-01-28 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS5
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 510A5 heavy chain, ... | | 著者 | Guo, H, Gao, Y, Ji, X, Yang, H. | | 登録日 | 2022-01-28 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS1
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 510A5 heavy chain, ... | | 著者 | Guo, H, Gao, Y, Ji, X, Yang, H. | | 登録日 | 2022-01-28 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
9CAY
 
 | |
8HUD
 
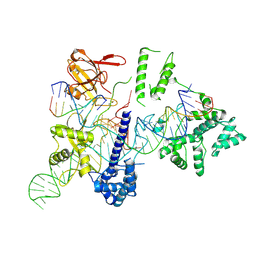 | | Cryo-EM structure of the EvCas9-sgRNA-target DNA ternary complex | | 分子名称: | CRISPR-associated endonuclease Cas9, Non-target DNA strand, Target DNA strand, ... | | 著者 | Tang, N, Wu, Z, Gao, Y, Chen, W, Su, M, Wang, Z, Ji, Q. | | 登録日 | 2022-12-23 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.43 Å) | | 主引用文献 | Molecular Basis and Genome Editing Applications of a Compact Eubacterium ventriosum CRISPR-Cas9 System.
Acs Synth Biol, 13, 2024
|
|
5XB0
 
 | |
7WWK
 
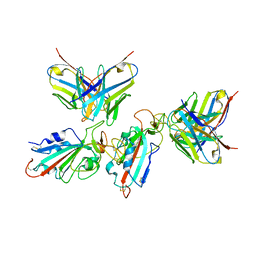 | | Local refinement of the SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab | | 分子名称: | 55A8 light chain, Spike glycoprotein | | 著者 | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | 登録日 | 2022-02-13 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Local refinement of the SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab
To Be Published
|
|
7WWJ
 
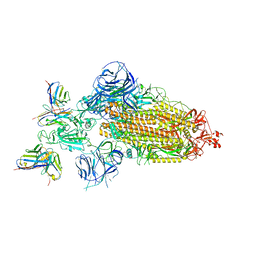 | | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab in the class 2 conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 55A8 heavy chain, ... | | 著者 | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | 登録日 | 2022-02-13 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | EM structure of SARS-CoV-2 Omicron variant spike glycoprotein and 55A8
To Be Published
|
|
7WWI
 
 | | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab in the class 1 conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 55A8 heavy chain, ... | | 著者 | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | 登録日 | 2022-02-13 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab in the class 1 conformation
To Be Published
|
|
7D5I
 
 | | Structure of Mycobacterium smegmatis bd complex in the apo-form. | | 分子名称: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Cytochrome D ubiquinol oxidase subunit 1, HEME B/C, ... | | 著者 | Wang, W, Gong, H, Gao, Y, Zhou, X, Rao, Z. | | 登録日 | 2020-09-26 | | 公開日 | 2021-06-23 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.79 Å) | | 主引用文献 | Cryo-EM structure of mycobacterial cytochrome bd reveals two oxygen access channels.
Nat Commun, 12, 2021
|
|
7XJ9
 
 | | Local refinement of the SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab | | 分子名称: | 55A8 heavy chain, 55A8 light chain, 58G6 heavy chain, ... | | 著者 | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | 登録日 | 2022-04-15 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | Local refinement of the SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab
To Be Published
|
|
7XJ8
 
 | | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab in the class 2 conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 55A8 heavy chain, ... | | 著者 | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | 登録日 | 2022-04-15 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab in the class 2 conformation
To Be Published
|
|
