4JX6
 
 | |
3EKZ
 
 | | Structural Characterization of tetrameric Mycobacterium tuberculosis fructose 1,6-bisphosphate aldolase - substrate binding and catalysis mechanism of a class IIa bacterial aldolase | | 分子名称: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-bisphosphate aldolase, SN-GLYCEROL-3-PHOSPHATE, ... | | 著者 | Pegan, S, Rukseree, K, Franzblau, S.G, Mesecar, A.D. | | 登録日 | 2008-09-19 | | 公開日 | 2009-02-10 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Structural basis for catalysis of a tetrameric class IIa fructose 1,6-bisphosphate aldolase from Mycobacterium tuberculosis
J.Mol.Biol., 386, 2009
|
|
6VGY
 
 | | 2.05 A resolution structure of MERS 3CL protease in complex with inhibitor 6b | | 分子名称: | N~2~-{[(trans-4-ethylcyclohexyl)oxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, Orf1a protein | | 著者 | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-01-09 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
6UXQ
 
 | | Crystal structure of BAK core domain BH3-groove-dimer in complex with POPC and C8E4 | | 分子名称: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-ETHANEDIOL, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, ... | | 著者 | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | 登録日 | 2019-11-07 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.696 Å) | | 主引用文献 | BAK core dimers bind lipids and can be bridged by them.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3EJ3
 
 | | Structural and mechanistic analysis of trans-3-chloroacrylic acid dehalogenase activity | | 分子名称: | ACETATE ION, Alpha-subunit of trans-3-chloroacrylic acid dehalogenase, Beta-subunit of trans-3-chloroacrylic acid dehalogenase, ... | | 著者 | Pegan, S, Serrano, H, Whitman, C.P, Mesecar, A.D. | | 登録日 | 2008-09-17 | | 公開日 | 2008-12-02 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and mechanistic analysis of trans-3-chloroacrylic acid dehalogenase activity.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
6UUG
 
 | | Structure of methanesulfinate monooxygenase MsuC from Pseudomonas fluorescens at 1.69 angstrom resolution | | 分子名称: | Putative dehydrogenase | | 著者 | Soule, J, Gnann, A.D, Gonzalez, R, Parker, M.J, McKenna, K.C, Nguyen, S.V, Phan, N.T, Wicht, D.K, Dowling, D.P. | | 登録日 | 2019-10-30 | | 公開日 | 2019-12-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.685 Å) | | 主引用文献 | Structure and function of the two-component flavin-dependent methanesulfinate monooxygenase within bacterial sulfur assimilation.
Biochem.Biophys.Res.Commun., 522, 2020
|
|
6VH3
 
 | | 2.20 A resolution structure of MERS 3CL protease in complex with inhibitor 7j | | 分子名称: | (1S,2S)-2-[(N-{[(4,4-difluorocyclohexyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, Orf1a protein | | 著者 | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-01-09 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
3EJ7
 
 | | Structural and mechanistic analysis of trans-3-chloroacrylic acid dehalogenase activity | | 分子名称: | Alpha-subunit of trans-3-chloroacrylic acid dehalogenase, Beta-subunit of trans-3-chloroacrylic acid dehalogenase, SULFATE ION | | 著者 | Pegan, S, Serrano, H, Whitman, C.P, Mesecar, A.D. | | 登録日 | 2008-09-17 | | 公開日 | 2008-12-02 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and mechanistic analysis of trans-3-chloroacrylic acid dehalogenase activity.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3ELF
 
 | | Structural Characterization of tetrameric Mycobacterium tuberculosis fructose 1,6-bisphosphate aldolase - substrate binding and catalysis mechanism of a class IIa bacterial aldolase | | 分子名称: | 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), Fructose-bisphosphate aldolase, SODIUM ION, ... | | 著者 | Pegan, S, Rukseree, K, Franzblau, S.G, Mesecar, A.D. | | 登録日 | 2008-09-22 | | 公開日 | 2009-02-10 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | Structural basis for catalysis of a tetrameric class IIa fructose 1,6-bisphosphate aldolase from Mycobacterium tuberculosis
J.Mol.Biol., 386, 2009
|
|
3EJ9
 
 | | Structural and mechanistic analysis of trans-3-chloroacrylic acid dehalogenase activity | | 分子名称: | Alpha-subunit of trans-3-chloroacrylic acid dehalogenase, Beta-subunit of trans-3-chloroacrylic acid dehalogenase | | 著者 | Pegan, S, Serrano, H, Whitman, C.P, Mesecar, A.D. | | 登録日 | 2008-09-17 | | 公開日 | 2008-12-02 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural and mechanistic analysis of trans-3-chloroacrylic acid dehalogenase activity.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3EKL
 
 | | Structural Characterization of tetrameric Mycobacterium tuberculosis fructose 1,6-bisphosphate aldolase - substrate binding and catalysis mechanism of a class IIa bacterial aldolase | | 分子名称: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-bisphosphate aldolase, SODIUM ION, ... | | 著者 | Pegan, S, Rukseree, K, Franzblau, S.G, Mesecar, A.D. | | 登録日 | 2008-09-19 | | 公開日 | 2009-02-10 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Structural basis for catalysis of a tetrameric class IIa fructose 1,6-bisphosphate aldolase from Mycobacterium tuberculosis
J.Mol.Biol., 386, 2009
|
|
6UXN
 
 | | Crystal structure of BAK core domain BH3-groove-dimer in complex with phosphatidylserine | | 分子名称: | Bcl-2 homologous antagonist/killer, GLYCEROL, O-[(R)-{[(2R)-2,3-bis(octanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, ... | | 著者 | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | 登録日 | 2019-11-07 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | BAK core dimers bind lipids and can be bridged by them.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6UXP
 
 | | Crystal structure of BAK core domain BH3-groove-dimer in complex with phosphatidylglycerol | | 分子名称: | 1,2-DIOCTANOYL-SN-GLYCERO-3-[PHOSPHO-RAC-(1-GLYCEROL), Bcl-2 homologous antagonist/killer, GLYCEROL | | 著者 | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | 登録日 | 2019-11-07 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.492 Å) | | 主引用文献 | BAK core dimers bind lipids and can be bridged by them.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VH2
 
 | | 2.26 A resolution structure of MERS 3CL protease in complex with inhibitor 7i | | 分子名称: | 4,4-difluorocyclohexyl [(2S)-3-cyclohexyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamate, Orf1a protein | | 著者 | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-01-09 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
6VH0
 
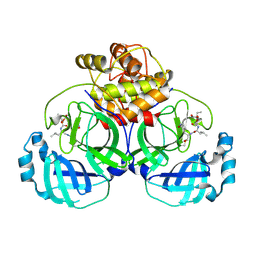 | | 1.95 A resolution structure of MERS 3CL protease in complex with inhibitor 6g | | 分子名称: | N~2~-{[(5-ethyl-1,3-dioxan-5-yl)oxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, Orf1a protein | | 著者 | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-01-09 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
6UXM
 
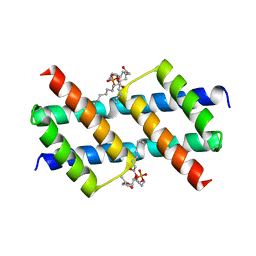 | |
6UXR
 
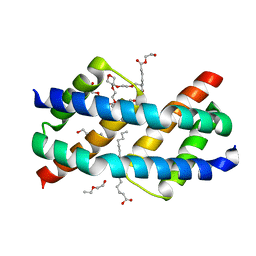 | | Crystal structure of BAK core domain BH3-groove-dimer in complex with LysoPC | | 分子名称: | Bcl-2 homologous antagonist/killer, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | 著者 | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | 登録日 | 2019-11-07 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | BAK core dimers bind lipids and can be bridged by them.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VH1
 
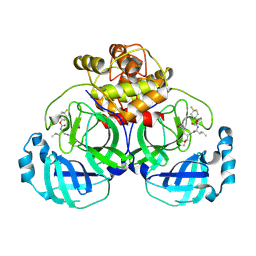 | | 2.30 A resolution structure of MERS 3CL protease in complex with inhibitor 6h | | 分子名称: | N~2~-{[(4,4-difluorocyclohexyl)oxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, Orf1a protein | | 著者 | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-01-09 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
6VGZ
 
 | | 2.25 A resolution structure of MERS 3CL protease in complex with inhibitor 6d | | 分子名称: | N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[trans-4-(propan-2-yl)cyclohexyl]oxy}carbonyl)-L-leucinamide, Orf1a protein | | 著者 | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-01-09 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
3D0X
 
 | |
6W5L
 
 | | 2.1 A resolution structure of Norovirus 3CL protease in complex with inhibitor 7g | | 分子名称: | (2~{S})-~{N}-[(1~{R})-1-[bis($l^{1}-oxidanyl)-methoxy-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{R})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-2-[[[2-(3-chlorophenyl)-2-methyl-propoxy]-oxidanylidene-methyl]amino]-4-methyl-pentanamide, 3C-LIKE PROTEASE | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-03-13 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
4M5W
 
 | | Crystal structure of the USP7/HAUSP catalytic domain | | 分子名称: | BROMIDE ION, Ubiquitin carboxyl-terminal hydrolase 7 | | 著者 | Molland, K.L, Mesecar, A.D, Zhou, Q. | | 登録日 | 2013-08-08 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.244 Å) | | 主引用文献 | A 2.2 angstrom resolution structure of the USP7 catalytic domain in a new space group elaborates upon structural rearrangements resulting from ubiquitin binding.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
6W5K
 
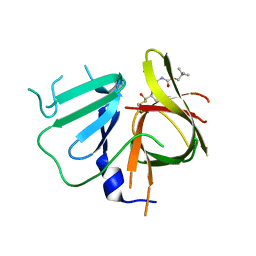 | | 1.95 A resolution structure of Norovirus 3CL protease in complex with inhibitor 5g | | 分子名称: | 3C-LIKE PROTEASE, N~2~-{[2-(3-chlorophenyl)-2-methylpropoxy]carbonyl}-N-{(1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}-L-leucinamide | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-03-13 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
6W81
 
 | |
4JX5
 
 | |
