6VQH
 
 | |
6VQ9
 
 | |
6VQ6
 
 | |
6VLS
 
 | | Structure of C-terminal fragment of Vip3A toxin | | 分子名称: | DI(HYDROXYETHYL)ETHER, Maltose/maltodextrin-binding periplasmic protein,Vip3Aa | | 著者 | Jiang, K, Zhang, Y, Chen, Z, Gao, X. | | 登録日 | 2020-01-25 | | 公開日 | 2020-07-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural and Functional Insights into the C-terminal Fragment of Insecticidal Vip3A Toxin ofBacillus thuringiensis.
Toxins, 12, 2020
|
|
6OQV
 
 | | E. coli ATP Synthase State 2b | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | 著者 | Stewart, A.G, Sobti, M, Walshe, J.L. | | 登録日 | 2019-04-29 | | 公開日 | 2020-06-03 | | 最終更新日 | 2025-05-21 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
3KDH
 
 | | Structure of ligand-free PYL2 | | 分子名称: | Putative uncharacterized protein At2g26040 | | 著者 | Yin, P, Fan, H, Hao, Q, Yuan, X, Yan, N. | | 登録日 | 2009-10-22 | | 公開日 | 2009-11-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.653 Å) | | 主引用文献 | Structural insights into the mechanism of abscisic acid signaling by PYL proteins
Nat.Struct.Mol.Biol., 16, 2009
|
|
3KDJ
 
 | | Complex structure of (+)-ABA-bound PYL1 and ABI1 | | 分子名称: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, MANGANESE (II) ION, Protein phosphatase 2C 56, ... | | 著者 | Yin, P, Fan, H, Hao, Q, Yuan, X, Yan, N. | | 登録日 | 2009-10-23 | | 公開日 | 2009-11-10 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.878 Å) | | 主引用文献 | Structural insights into the mechanism of abscisic acid signaling by PYL proteins
Nat.Struct.Mol.Biol., 16, 2009
|
|
5E3S
 
 | |
5E3U
 
 | | Crystal structure of phosphatidylinositol-4-phosphate 5-kinase | | 分子名称: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Phosphatidylinositol-4-phosphate 5-kinase, ... | | 著者 | Muftuoglu, Y. | | 登録日 | 2015-10-04 | | 公開日 | 2016-07-20 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Mechanism of substrate specificity of phosphatidylinositol phosphate kinases.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5E3T
 
 | | Crystal structure of phosphatidylinositol-4-phosphate 5-kinase | | 分子名称: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Phosphatidylinositol-4-phosphate 5-kinase, ... | | 著者 | Muftuoglu, Y. | | 登録日 | 2015-10-04 | | 公開日 | 2016-07-20 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Mechanism of substrate specificity of phosphatidylinositol phosphate kinases.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7WD3
 
 | | Cryo-EM structure of Drg1 hexamer treated with ATP and benzo-diazaborine | | 分子名称: | 2-(TOLUENE-4-SULFONYL)-2H-BENZO[D][1,2,3]DIAZABORININ-1-OL, ADENOSINE-5'-TRIPHOSPHATE, ATPase family gene 2 protein | | 著者 | Ma, C.Y, Wu, D.M, Chen, Q, Gao, N. | | 登録日 | 2021-12-21 | | 公開日 | 2022-12-14 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition.
Nat Commun, 13, 2022
|
|
7WBB
 
 | | Cryo-EM structure of substrate engaged Drg1 hexamer | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, AFG2 isoform 1, substrate | | 著者 | Ma, C.Y, Wu, D.M, Chen, Q, Gao, N. | | 登録日 | 2021-12-16 | | 公開日 | 2022-12-28 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition.
Nat Commun, 13, 2022
|
|
3KDI
 
 | | Structure of (+)-ABA bound PYL2 | | 分子名称: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Putative uncharacterized protein At2g26040 | | 著者 | Yin, P, Fan, H, Hao, Q, Yuan, X, Yan, N. | | 登録日 | 2009-10-22 | | 公開日 | 2009-11-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.379 Å) | | 主引用文献 | Structural insights into the mechanism of abscisic acid signaling by PYL proteins
Nat.Struct.Mol.Biol., 16, 2009
|
|
1FSH
 
 | |
3GMQ
 
 | | Structure of mouse CD1d expressed in SF9 cells, no ligand added | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2 microglobulin, ... | | 著者 | Schiefner, A, Wilson, I.A. | | 登録日 | 2009-03-14 | | 公開日 | 2009-11-10 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural evaluation of potent NKT cell agonists: implications for design of novel stimulatory ligands.
J.Mol.Biol., 394, 2009
|
|
8IED
 
 | | Cryo-EM structure of GPR156-miniGo-scFv16 complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(o) subunit alpha, ... | | 著者 | Shin, J, Park, J, Cho, Y. | | 登録日 | 2023-02-15 | | 公開日 | 2024-02-14 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.33 Å) | | 主引用文献 | Constitutive activation mechanism of a class C GPCR.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8IEI
 
 | |
8IEQ
 
 | | Cryo-EM structure of G-protein free GPR156 | | 分子名称: | Probable G-protein coupled receptor 156, [(2R)-3-[(E)-hexadec-9-enoyl]oxy-2-octadecanoyloxy-propyl] 2-(trimethylazaniumyl)ethyl phosphate | | 著者 | Shin, J, Park, J, Cho, Y. | | 登録日 | 2023-02-15 | | 公開日 | 2024-02-14 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (2.73 Å) | | 主引用文献 | Constitutive activation mechanism of a class C GPCR.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8JP2
 
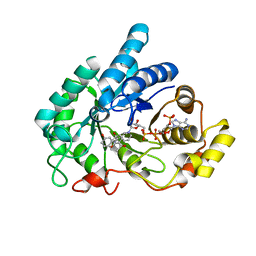 | | Crystal structure of AKR1C1 in complex with DFV | | 分子名称: | 7-HYDROXY-2-(4-HYDROXY-PHENYL)-CHROMAN-4-ONE, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Zheng, X.H, Liu, H, Yao, Z.Q, Zhang, L.P. | | 登録日 | 2023-06-10 | | 公開日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Inhibition of AKR1Cs by liquiritigenin and the structural basis.
Chem.Biol.Interact., 385, 2023
|
|
8JP1
 
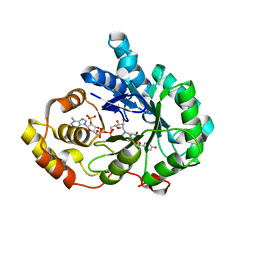 | | Crystal structure of AKR1C3 in complex with DFV | | 分子名称: | 7-HYDROXY-2-(4-HYDROXY-PHENYL)-CHROMAN-4-ONE, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Zheng, X.H, Liu, H, Yao, Z.Q, Zhang, L.P. | | 登録日 | 2023-06-10 | | 公開日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Inhibition of AKR1Cs by liquiritigenin and the structural basis.
Chem.Biol.Interact., 385, 2023
|
|
8IEC
 
 | | Cryo-EM structure of miniGo-scFv16 of GPR156-miniGo-scFv16 complex (local refine) | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(o) subunit alpha, ... | | 著者 | Shin, J, Park, J, Cho, Y. | | 登録日 | 2023-02-15 | | 公開日 | 2024-02-14 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Constitutive activation mechanism of a class C GPCR.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8IEB
 
 | |
8IEP
 
 | |
3GMP
 
 | | Structure of mouse CD1d in complex with PBS-25 | | 分子名称: | (2S,3S,4R)-N-OCTANOYL-1-[(ALPHA-D-GALACTOPYRANOSYL)OXY]-2-AMINO-OCTADECANE-3,4-DIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Schiefner, A, Wilson, I.A. | | 登録日 | 2009-03-14 | | 公開日 | 2009-11-10 | | 最終更新日 | 2024-11-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural evaluation of potent NKT cell agonists: implications for design of novel stimulatory ligands.
J.Mol.Biol., 394, 2009
|
|
3GML
 
 | | Structure of mouse CD1d in complex with C6Ph | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2 microglobulin, ... | | 著者 | Schiefner, A, Wilson, I.A. | | 登録日 | 2009-03-14 | | 公開日 | 2009-11-10 | | 最終更新日 | 2024-11-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural evaluation of potent NKT cell agonists: implications for design of novel stimulatory ligands.
J.Mol.Biol., 394, 2009
|
|
