4LEN
 
 | |
4M51
 
 | | Crystal structure of amidohydrolase nis_0429 (ser145ala mutant) from nitratiruptor sp. sb155-2 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Amidohydrolase family protein, BENZOIC ACID, ... | | 著者 | Patskovsky, Y, Toro, R, Gobble, A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-08-07 | | 公開日 | 2013-09-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | Deamination of 6-aminodeoxyfutalosine in menaquinone biosynthesis by distantly related enzymes.
Biochemistry, 52, 2013
|
|
4JN0
 
 | |
4JMS
 
 | |
4JQN
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 4-Hydroxybenzaldehyde | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cytochrome c peroxidase, P-HYDROXYBENZALDEHYDE, ... | | 著者 | Boyce, S.E, Fischer, M, Fish, I. | | 登録日 | 2013-03-20 | | 公開日 | 2013-07-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
4JQJ
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 4-Aminoquinoline | | 分子名称: | Cytochrome c peroxidase, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Boyce, S.E, Fischer, M, Fish, I. | | 登録日 | 2013-03-20 | | 公開日 | 2013-07-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
4JMV
 
 | |
4M8T
 
 | |
3HPA
 
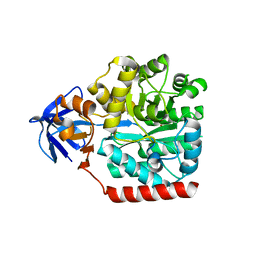 | | Crystal structure of an amidohydrolase gi:44264246 from an evironmental sample of sargasso sea | | 分子名称: | AMIDOHYDROLASE, ZINC ION | | 著者 | Fedorov, A.A, Fedorov, E.V, Toro, R, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-06-03 | | 公開日 | 2009-06-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The hunt for 8-oxoguanine deaminase.
J.Am.Chem.Soc., 132, 2010
|
|
3ID7
 
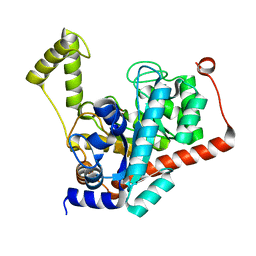 | | Crystal structure of renal dipeptidase from Streptomyces coelicolor A3(2) | | 分子名称: | CHLORIDE ION, Dipeptidase, ZINC ION | | 著者 | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | 登録日 | 2009-07-20 | | 公開日 | 2010-01-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structure, mechanism, and substrate profile for Sco3058: the closest bacterial homologue to human renal dipeptidase .
Biochemistry, 49, 2010
|
|
3FDK
 
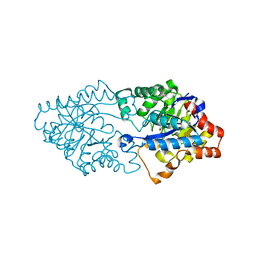 | | Crystal structure of hydrolase DR0930 with promiscuous catalytic activity | | 分子名称: | HYDROLASE DR0930, ZINC ION | | 著者 | Fedorov, A.A, Fedorov, L.V, Xiang, D.F, Raushel, F.M, Almo, S.C. | | 登録日 | 2008-11-25 | | 公開日 | 2009-06-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Functional annotation and three-dimensional structure of Dr0930 from Deinococcus radiodurans, a close relative of phosphotriesterase in the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3ITC
 
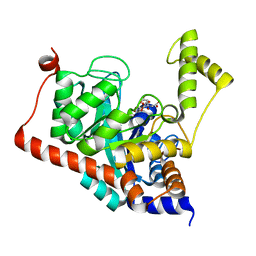 | | Crystal structure of Sco3058 with bound citrate and glycerol | | 分子名称: | CITRIC ACID, GLYCEROL, ZINC ION, ... | | 著者 | Nguyen, T.T, Cummings, J.A, Tsai, C.-L, Barondeau, D.P, Raushel, F.M. | | 登録日 | 2009-08-28 | | 公開日 | 2010-02-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure, mechanism, and substrate profile for Sco3058: the closest bacterial homologue to human renal dipeptidase
Biochemistry, 49, 2010
|
|
2AAZ
 
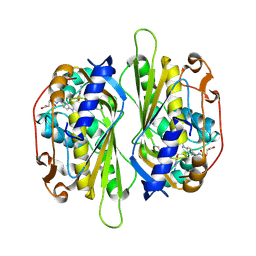 | | Cryptococcus neoformans thymidylate synthase complexed with substrate and an antifolate | | 分子名称: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | 著者 | Finer-Moore, J.S, Anderson, A.C, O'Neil, R.H, Costi, M.P, Ferrari, S, Krucinski, J, Stroud, R.M. | | 登録日 | 2005-07-14 | | 公開日 | 2005-12-06 | | 最終更新日 | 2018-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | The structure of Cryptococcus neoformans thymidylate synthase suggests strategies for using target dynamics for species-specific inhibition.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5SQ0
 
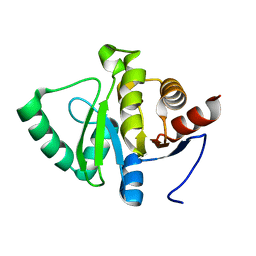 | |
5SPZ
 
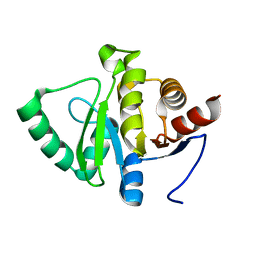 | |
5SPX
 
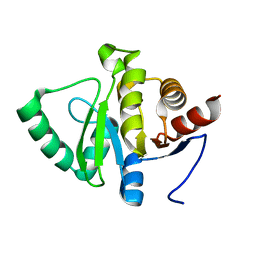 | |
5SQ1
 
 | |
5SQ2
 
 | |
5SQ3
 
 | |
5SPY
 
 | |
5SQ4
 
 | |
5SQ8
 
 | |
5SQB
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894390 - (R,R) and (S,S) isomers | | 分子名称: | (1R,2R)-1-({6-[(cyclopropylmethyl)amino]pyridine-3-carbonyl}amino)-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-1-({6-[(cyclopropylmethyl)amino]pyridine-3-carbonyl}amino)-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-06-09 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SQC
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894388 - (R,R) and (S,S) isomers | | 分子名称: | (1R,2R)-4-hydroxy-1-({5-[(oxan-4-yl)amino]pyrazine-2-carbonyl}amino)-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-4-hydroxy-1-({5-[(oxan-4-yl)amino]pyrazine-2-carbonyl}amino)-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-06-09 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SQ6
 
 | |
