7VSC
 
 | | E. coli Ribonuclease HI in complex with one Mg2+ (1) | | 分子名称: | MAGNESIUM ION, Ribonuclease HI, SULFATE ION | | 著者 | Liao, Z, Oyama, T. | | 登録日 | 2021-10-26 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Pivotal role of a conserved histidine in Escherichia coli ribonuclease HI as proposed by X-ray crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7VSD
 
 | | E. coli Ribonuclease HI in complex with one Mg2+ (2) | | 分子名称: | MAGNESIUM ION, Ribonuclease HI, SULFATE ION | | 著者 | Liao, Z, Oyama, T. | | 登録日 | 2021-10-26 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Pivotal role of a conserved histidine in Escherichia coli ribonuclease HI as proposed by X-ray crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7VSE
 
 | |
7W51
 
 | |
7W4Z
 
 | |
7W50
 
 | |
7W52
 
 | |
5WVO
 
 | | Crystal structure of DNMT1 RFTS domain in complex with K18/K23 mono-ubiquitylated histone H3 | | 分子名称: | DNA (cytosine-5)-methyltransferase 1, Histone H3.1, Ubiquitin, ... | | 著者 | Ishiyama, S, Nishiyama, A, Nakanishi, M, Arita, K. | | 登録日 | 2016-12-28 | | 公開日 | 2017-11-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.997 Å) | | 主引用文献 | Structure of the Dnmt1 Reader Module Complexed with a Unique Two-Mono-Ubiquitin Mark on Histone H3 Reveals the Basis for DNA Methylation Maintenance
Mol. Cell, 68, 2017
|
|
1LHM
 
 | |
2YJ8
 
 | | CATHEPSIN L WITH A NITRILE INHIBITOR | | 分子名称: | (2S,4R)-4-(2-chlorophenyl)sulfonyl-N-[1-(iminomethyl)cyclopropyl]-1-[1-(4-iodophenyl)cyclopropyl]carbonyl-pyrrolidine-2-carboxamide, CATHEPSIN L1, GLYCEROL | | 著者 | Banner, D.W, Benz, J.M, Haap, W. | | 登録日 | 2011-05-19 | | 公開日 | 2011-11-23 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Halogen Bonding at the Active Sites of Human Cathepsin L and Mek1 Kinase: Efficient Interactions in Different Environments.
Chemmedchem, 6, 2011
|
|
2YJB
 
 | | CATHEPSIN L WITH A NITRILE INHIBITOR | | 分子名称: | (2S,4R)-4-(2-chlorophenyl)sulfonyl-N-[1-(iminomethyl)cyclopropyl]-1-[1-[4-(trifluoromethyl)phenyl]cyclopropyl]carbonyl-pyrrolidine-2-carboxamide, CATHEPSIN L1, GLYCEROL | | 著者 | Banner, D.W, Benz, J.M, Haap, W. | | 登録日 | 2011-05-19 | | 公開日 | 2011-11-23 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Halogen Bonding at the Active Sites of Human Cathepsin L and Mek1 Kinase: Efficient Interactions in Different Environments.
Chemmedchem, 6, 2011
|
|
7YNE
 
 | |
2YYW
 
 | |
3VYL
 
 | | Structure of L-ribulose 3-epimerase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, L-ribulose 3-epimerase, MANGANESE (II) ION | | 著者 | Uechi, K, Sakuraba, H, Takata, G. | | 登録日 | 2012-09-27 | | 公開日 | 2013-10-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural insight into L-ribulose 3-epimerase from Mesorhizobium loti.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2YYX
 
 | |
2ZFU
 
 | | Structure of the methyltransferase-like domain of nucleomethylin | | 分子名称: | Cerebral protein 1, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Minami, H, Hashimoto, H, Murayama, A, Yanagisawa, J, Sato, M, Shimizu, T. | | 登録日 | 2008-01-14 | | 公開日 | 2008-12-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Epigenetic control of rDNA loci in response to intracellular energy status
Cell(Cambridge,Mass.), 133, 2008
|
|
5V7T
 
 | | crystal structure of PARP14 bound to N-{4-[4-(diphenylmethoxy)piperidin-1-yl]butyl}[1,2,4]triazolo[4,3-b]pyridazin-6-amine inhibitor | | 分子名称: | N-{4-[4-(diphenylmethoxy)piperidin-1-yl]butyl}[1,2,4]triazolo[4,3-b]pyridazin-6-amine, Poly [ADP-ribose] polymerase 14 | | 著者 | saikatendu, k.s, Hirozane, M. | | 登録日 | 2017-03-20 | | 公開日 | 2017-05-03 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Identification of PARP14 inhibitors using novel methods for detecting auto-ribosylation.
Biochem. Biophys. Res. Commun., 486, 2017
|
|
5V7W
 
 | | Crystal structure of human PARP14 bound to 2-{[(1-methylpiperidin-4-yl)methyl]amino}-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4(3H)-one inhibitor | | 分子名称: | 2-{[(1-methylpiperidin-4-yl)methyl]amino}-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4(3H)-one, Poly [ADP-ribose] polymerase 14 | | 著者 | saikatendu, k.s, Hirozane, M. | | 登録日 | 2017-03-20 | | 公開日 | 2017-05-10 | | 最終更新日 | 2018-11-14 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Identification of PARP14 inhibitors using novel methods for detecting auto-ribosylation.
Biochem. Biophys. Res. Commun., 486, 2017
|
|
2Z47
 
 | |
3WW3
 
 | | X-ray structures of Cellulomonas parahominis L-ribose isomerase with no ligand | | 分子名称: | L-ribose isomerase, MANGANESE (II) ION | | 著者 | Terami, Y, Yoshida, H, Takata, G, Kamitori, S. | | 登録日 | 2014-06-13 | | 公開日 | 2015-04-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Essentiality of tetramer formation of Cellulomonas parahominis L-ribose isomerase involved in novel L-ribose metabolic pathway.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3WW1
 
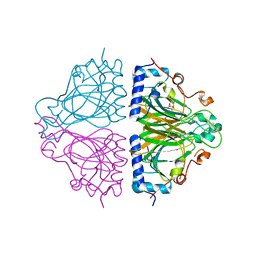 | | X-ray structure of Cellulomonas parahominis L-ribose isomerase with L-ribose | | 分子名称: | L-ribose, L-ribose isomerase, MANGANESE (II) ION, ... | | 著者 | Terami, Y, Yoshida, H, Takata, G, Kamitori, S. | | 登録日 | 2014-06-13 | | 公開日 | 2015-04-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Essentiality of tetramer formation of Cellulomonas parahominis L-ribose isomerase involved in novel L-ribose metabolic pathway.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3WW4
 
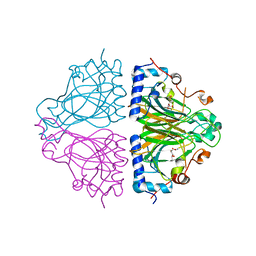 | | X-ray structures of Cellulomonas parahominis L-ribose isomerase with L-allose | | 分子名称: | L-allose, L-ribose isomerase, MANGANESE (II) ION, ... | | 著者 | Terami, Y, Yoshida, H, Takata, G, Kamitori, S. | | 登録日 | 2014-06-13 | | 公開日 | 2015-04-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Essentiality of tetramer formation of Cellulomonas parahominis L-ribose isomerase involved in novel L-ribose metabolic pathway.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3WIG
 
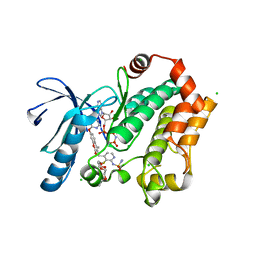 | | Human MEK1 kinase in complex with CH5126766 and MgAMP-PNP | | 分子名称: | CHLORIDE ION, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | 著者 | Lukacs, C.M, Janson, C, Schuck, V. | | 登録日 | 2013-09-12 | | 公開日 | 2014-06-04 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Disruption of CRAF-Mediated MEK Activation Is Required for Effective MEK Inhibition in KRAS Mutant Tumors
Cancer Cell, 25, 2014
|
|
3WW2
 
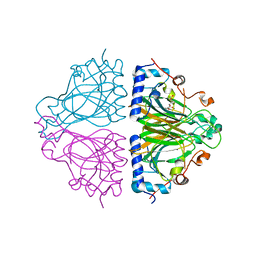 | | X-ray structures of Cellulomonas parahominis L-ribose isomerase with L-psicose | | 分子名称: | L-psicose, L-ribose isomerase, MANGANESE (II) ION, ... | | 著者 | Terami, Y, Yoshida, H, Takata, G, Kamitori, S. | | 登録日 | 2014-06-13 | | 公開日 | 2015-04-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Essentiality of tetramer formation of Cellulomonas parahominis L-ribose isomerase involved in novel L-ribose metabolic pathway.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3B12
 
 | |
