8D1H
 
 | | hBest2 Ca2+-unbound closed state | | 分子名称: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, Bestrophin-2, ... | | 著者 | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | 登録日 | 2022-05-27 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (1.94 Å) | | 主引用文献 | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
4Y5U
 
 | | Transcription factor | | 分子名称: | NICKEL (II) ION, Signal transducer and activator of transcription 6 | | 著者 | Li, J, Niu, F, Ouyang, S, Liu, Z. | | 登録日 | 2015-02-12 | | 公開日 | 2016-02-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.708 Å) | | 主引用文献 | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4Y5W
 
 | | Transcription factor-DNA complex | | 分子名称: | DNA (5'-D(P*AP*TP*GP*GP*AP*TP*TP*TP*CP*CP*TP*AP*GP*GP*AP*AP*GP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*CP*TP*TP*CP*CP*TP*AP*GP*GP*AP*AP*AP*TP*CP*CP*AP*T)-3'), Signal transducer and activator of transcription 6 | | 著者 | Li, J, Niu, F, Ouyang, S, Liu, Z. | | 登録日 | 2015-02-12 | | 公開日 | 2016-02-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.104 Å) | | 主引用文献 | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8W5J
 
 | | Cryo-EM structure of the yeast TOM core complex (from TOM-TIM23 complex) | | 分子名称: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | 著者 | Wang, Q, Guan, Z.Y, Zhuang, J.J, Huang, R, Yin, P. | | 登録日 | 2023-08-27 | | 公開日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | The architecture of substrate-engaged TOM-TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation.
Cell Discov, 10, 2024
|
|
7OJ1
 
 | |
7OJ2
 
 | | Bacillus subtilis IMPDH in complex with Ap4A | | 分子名称: | GLYCEROL, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, PHOSPHATE ION | | 著者 | Giammarinaro, P.I, Bange, G. | | 登録日 | 2021-05-13 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Diadenosine tetraphosphate regulates biosynthesis of GTP in Bacillus subtilis.
Nat Microbiol, 7, 2022
|
|
7D3I
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with MI-23 | | 分子名称: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[3-[3,5-bis(fluoranyl)phenyl]propanoyl]-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase | | 著者 | Zeng, R, Li, Y.S, Qiao, J.X, Wang, Y.F, Yang, S.Y, Lei, J. | | 登録日 | 2020-09-19 | | 公開日 | 2020-10-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.004 Å) | | 主引用文献 | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
8ECY
 
 | | cryoEM structure of bovine bestrophin-2 and glutamine synthetase complex | | 分子名称: | Bestrophin, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Owji, A.P, Kittredge, A.K, Yang, T. | | 登録日 | 2022-09-02 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2 Å) | | 主引用文献 | Bestrophin-2 and glutamine synthetase form a complex for glutamate release.
Nature, 611, 2022
|
|
7F46
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab (state1, local refinement of the RBD, NTD and 35B5 Fab) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | 著者 | Wang, X.F, Zhu, Y.Q. | | 登録日 | 2021-06-17 | | 公開日 | 2022-03-23 | | 最終更新日 | 2022-09-21 | | 実験手法 | ELECTRON MICROSCOPY (4.79 Å) | | 主引用文献 | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
5DAJ
 
 | | Crystal structure of NalD, the secondary repressor of MexAB-OprM multidrug efflux pump in Pseudomonas aeruginosa | | 分子名称: | NalD | | 著者 | Chen, W.Z, Wang, D, Huang, S.Q, Hu, Q.Y, Liu, X.C, Gan, J.H, Chen, H. | | 登録日 | 2015-08-20 | | 公開日 | 2016-04-20 | | 最終更新日 | 2023-04-12 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Novobiocin binding to NalD induces the expression of the MexAB-OprM pump in Pseudomonas aeruginosa
Mol.Microbiol., 100, 2016
|
|
7FAH
 
 | | Immune complex of head region of CA09 HA and neutralizing antibody 12H5 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | 著者 | Li, T.T, Xue, W.H, Gu, Y, Li, S.W. | | 登録日 | 2021-07-06 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.151 Å) | | 主引用文献 | Identification of a cross-neutralizing antibody that targets the receptor binding site of H1N1 and H5N1 influenza viruses.
Nat Commun, 13, 2022
|
|
6DL2
 
 | | BRD4 bromodomain 1 in complex with HYB157 | | 分子名称: | 1,2-ETHANEDIOL, 3-benzyl-2,9-dimethyl-4H,6H-thieno[2,3-e][1,2,4]triazolo[3,4-c][1,4]oxazepine, Bromodomain-containing protein 4 | | 著者 | Meagher, J.L, Stuckey, J.A. | | 登録日 | 2018-05-31 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Discovery of QCA570 as an Exceptionally Potent and Efficacious Proteolysis Targeting Chimera (PROTAC) Degrader of the Bromodomain and Extra-Terminal (BET) Proteins Capable of Inducing Complete and Durable Tumor Regression.
J. Med. Chem., 61, 2018
|
|
6LGN
 
 | | The atomic structure of varicella zoster virus C-capsid | | 分子名称: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | 著者 | Li, S, Zheng, Q. | | 登録日 | 2019-12-05 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (5.3 Å) | | 主引用文献 | Near-atomic cryo-electron microscopy structures of varicella-zoster virus capsids.
Nat Microbiol, 5, 2020
|
|
6INH
 
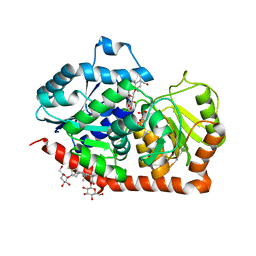 | | A glycosyltransferase with UDP and the substrate | | 分子名称: | 1-O-[(8alpha,9beta,10alpha,13alpha)-13-(beta-D-glucopyranosyloxy)-18-oxokaur-16-en-18-yl]-beta-D-glucopyranose, GLYCEROL, UDP-glycosyltransferase 76G1, ... | | 著者 | Zhu, X. | | 登録日 | 2018-10-25 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations.
Nat Commun, 10, 2019
|
|
2Y1R
 
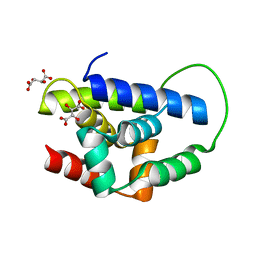 | | Structure of MecA121 & ClpC N-domain complex | | 分子名称: | ADAPTER PROTEIN MECA 1, NEGATIVE REGULATOR OF GENETIC COMPETENCE CLPC/MECB, S,R MESO-TARTARIC ACID | | 著者 | Wang, F, Mei, Z.Q, Wang, J.W, Shi, Y.G. | | 登録日 | 2010-12-10 | | 公開日 | 2011-03-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.595 Å) | | 主引用文献 | Structure and Mechanism of the Hexameric Meca-Clpc Molecular Machine.
Nature, 471, 2011
|
|
8F4O
 
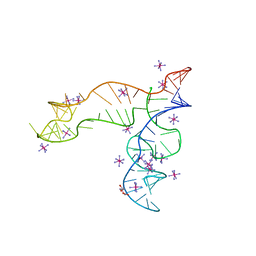 | | Apo structure of the TPP riboswitch aptamer domain | | 分子名称: | IRIDIUM HEXAMMINE ION, TETRAETHYLENE GLYCOL, TPP riboswitch aptamer domain, ... | | 著者 | Lee, H.-K, Wang, Y.-X, Stagno, J.R. | | 登録日 | 2022-11-11 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal structure of Escherichia coli thiamine pyrophosphate-sensing riboswitch in the apo state.
Structure, 31, 2023
|
|
6LGL
 
 | | The atomic structure of varicella-zoster virus A-capsid | | 分子名称: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | 著者 | Zheng, Q, Li, S. | | 登録日 | 2019-12-05 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Near-atomic cryo-electron microscopy structures of varicella-zoster virus capsids.
Nat Microbiol, 5, 2020
|
|
2Y1Q
 
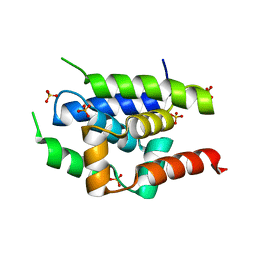 | | Crystal Structure of ClpC N-terminal Domain | | 分子名称: | NEGATIVE REGULATOR OF GENETIC COMPETENCE CLPC/MECB, SULFATE ION | | 著者 | Wang, F, Mei, Z.Q, Wang, J.W, Shi, Y.G. | | 登録日 | 2010-12-10 | | 公開日 | 2011-03-16 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure and Mechanism of the Hexameric Meca-Clpc Molecular Machine.
Nature, 471, 2011
|
|
7YR2
 
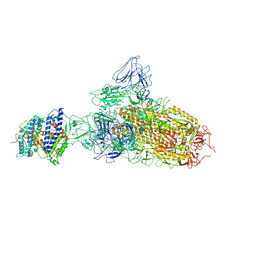 | | SARS-CoV-2 BA.2.75 S Trimer in complex with ACE2(state1) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Wang, L. | | 登録日 | 2022-08-08 | | 公開日 | 2022-10-19 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQU
 
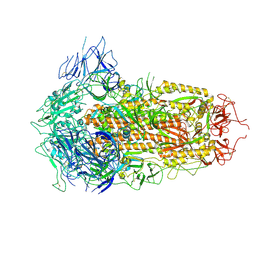 | | SARS-CoV-2 BA.2.75 S Trimer (3 RBD Down) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Wang, L. | | 登録日 | 2022-08-08 | | 公開日 | 2022-10-19 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.19 Å) | | 主引用文献 | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQZ
 
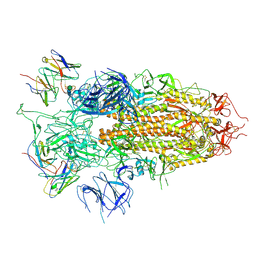 | | SARS-CoV-2 BA.2.75 S Trimer in complex with S309 (state3) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S309 heavy chain, S309 light chain, ... | | 著者 | Wang, L. | | 登録日 | 2022-08-08 | | 公開日 | 2022-10-19 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.84 Å) | | 主引用文献 | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YR0
 
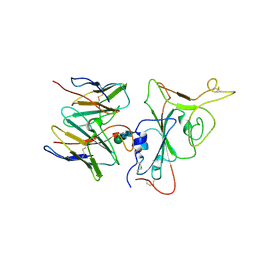 | |
7YQW
 
 | | SARS-CoV-2 BA.2.75 S Trimer (3 RBD Down) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Wang, L. | | 登録日 | 2022-08-08 | | 公開日 | 2022-10-19 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.51 Å) | | 主引用文献 | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQX
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with S309 (state1) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 heavy chain, ... | | 著者 | Wang, L. | | 登録日 | 2022-08-08 | | 公開日 | 2022-10-19 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.72 Å) | | 主引用文献 | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQY
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with S309 (state2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 heavy chain, ... | | 著者 | Wang, L. | | 登録日 | 2022-08-08 | | 公開日 | 2022-10-19 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.74 Å) | | 主引用文献 | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
