4ZAG
 
 | |
5LCN
 
 | |
1ADU
 
 | | EARLY E2A DNA-BINDING PROTEIN | | 分子名称: | ADENOVIRUS SINGLE-STRANDED DNA-BINDING PROTEIN, ZINC ION | | 著者 | Tucker, P.A, Kanellopoulos, P.N, Tsernoglou, D, Van Der Vliet, P.C. | | 登録日 | 1995-05-11 | | 公開日 | 1996-06-20 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Alternative arrangements of the protein chain are possible for the adenovirus single-stranded DNA binding protein.
J.Mol.Biol., 257, 1996
|
|
4ZAZ
 
 | | Structure of UbiX Y169F in complex with a covalent adduct formed between reduced FMN and dimethylallyl monophosphate | | 分子名称: | 1-deoxy-1-[7,8-dimethyl-5-(3-methylbut-2-en-1-yl)-2,4-dioxo-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-5-O-phosphono -D-ribitol, PHOSPHATE ION, SODIUM ION, ... | | 著者 | White, M.D, Leys, D. | | 登録日 | 2015-04-14 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis.
Nature, 522, 2015
|
|
1A3B
 
 | | COMPLEX OF HUMAN ALPHA-THROMBIN WITH THE BIFUNCTIONAL BORONATE INHIBITOR BOROLOG1 | | 分子名称: | ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), Hirudin, ... | | 著者 | Skordalakes, E, Elgendy, S, Dodson, G, Goodwin, C.A, Green, D, Scully, M.F, Freyssinet, J.H, Kakkar, V.V, Deadman, J. | | 登録日 | 1998-01-20 | | 公開日 | 1998-06-03 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Bifunctional peptide boronate inhibitors of thrombin: crystallographic analysis of inhibition enhanced by linkage to an exosite 1 binding peptide.
Biochemistry, 37, 1998
|
|
5LJN
 
 | | Structure of the HOIP PUB domain bound to SPATA2 PIM peptide | | 分子名称: | E3 ubiquitin-protein ligase RNF31, GLYCEROL, SULFATE ION, ... | | 著者 | Elliott, P.R, Komander, D. | | 登録日 | 2016-07-18 | | 公開日 | 2016-08-24 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.701 Å) | | 主引用文献 | SPATA2 Links CYLD to LUBAC, Activates CYLD, and Controls LUBAC Signaling.
Mol.Cell, 63, 2016
|
|
7TZO
 
 | | The apo structure of human mTORC2 complex | | 分子名称: | Rapamycin-insensitive companion of mTOR, Serine/threonine-protein kinase mTOR, Target of rapamycin complex 2 subunit MAPKAP1, ... | | 著者 | Yu, Z, Chen, J, Pearce, D. | | 登録日 | 2022-02-16 | | 公開日 | 2023-01-11 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | Interactions between mTORC2 core subunits Rictor and mSin1 dictate selective and context-dependent phosphorylation of substrate kinases SGK1 and Akt.
J.Biol.Chem., 298, 2022
|
|
5LMD
 
 | | The crystal structure of hCA II in complex with a benzoxaborole inhibitor | | 分子名称: | 1-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-4-yl]-3-(2-methoxy-5-methyl-phenyl)urea, Carbonic anhydrase 2, ZINC ION | | 著者 | De Simone, G, Alterio, V, Esposito, D, Di Fiore, A. | | 登録日 | 2016-07-29 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Benzoxaborole as a new chemotype for carbonic anhydrase inhibition.
Chem.Commun.(Camb.), 52, 2016
|
|
3UUO
 
 | | The discovery of potent, selectivity, and orally bioavailable pyrozoloquinolines as PDE10 inhibitors for the treatment of Schizophrenia | | 分子名称: | 6-methoxy-3,8-dimethyl-4-(piperazin-1-yl)-1H-pyrazolo[3,4-b]quinoline, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Ho, G.D, Yang, S, Smotryski, J, Bercovici, A, Nechuta, T, Smith, E.M, McElroy, W, Tan, Z, Tulshian, D, Mckittrick, B, Greenlee, W.J, Hruza, A, Xiao, L, Rindgen, D, Guzzi, M, Zhang, X, Bleickardt, C, Mullins, D, Hodgson, R. | | 登録日 | 2011-11-28 | | 公開日 | 2012-01-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | The discovery of potent, selective, and orally active pyrazoloquinolines as PDE10A inhibitors for the treatment of Schizophrenia.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5LOC
 
 | |
1F4V
 
 | | CRYSTAL STRUCTURE OF ACTIVATED CHEY BOUND TO THE N-TERMINUS OF FLIM | | 分子名称: | BERYLLIUM TRIFLUORIDE ION, CHEMOTAXIS CHEY PROTEIN, FLAGELLAR MOTOR SWITCH PROTEIN, ... | | 著者 | Lee, S.Y, Cho, H.S, Pelton, J.G, Yan, D, Henderson, R.K, King, D, Huang, L.S, Kustu, S, Berry, E.A, Wemmer, D.E. | | 登録日 | 2000-06-10 | | 公開日 | 2001-01-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Crystal structure of an activated response regulator bound to its target.
Nat.Struct.Biol., 8, 2001
|
|
2J50
 
 | | Structure of Aurora-2 in complex with PHA-739358 | | 分子名称: | N-[(3E)-5-[(2R)-2-METHOXY-2-PHENYLACETYL]PYRROLO[3,4-C]PYRAZOL-3(5H)-YLIDENE]-4-(4-METHYLPIPERAZIN-1-YL)BENZAMIDE, SERINE/THREONINE-PROTEIN KINASE 6, SULFATE ION | | 著者 | Cameron, A.D, Izzo, G, Storici, P, Rusconi, L, Fancelli, D, Varasi, M, Berta, D, Bindi, S, Forte, B, Severino, D, Tonani, R, Vianello, P. | | 登録日 | 2006-09-08 | | 公開日 | 2006-11-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | 1,4,5,6-Tetrahydropyrrolo[3,4-C]Pyrazoles: Identification of a Potent Aurora Kinase Inhibitor with a Favorable Antitumor Kinase Inhibition Profile.
J.Med.Chem., 49, 2006
|
|
3HLW
 
 | | CTX-M-9 S70G in complex with cefotaxime | | 分子名称: | (6R,7R)-3-(acetyloxymethyl)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyimino-ethanoyl]amino]-8-oxo-5-thia-1-azabicy clo[4.2.0]oct-2-ene-2-carboxylic acid, CTX-M-9 extended-spectrum beta-lactamase | | 著者 | Delmas, J, Leyssne, D, Dubois, D, Vazeille, E, Robin, F, Bonnet, R. | | 登録日 | 2009-05-28 | | 公開日 | 2010-06-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural insights into substrate recognition and product expulsion in CTX-M enzymes.
J.Mol.Biol., 400, 2010
|
|
8CYA
 
 | | SARS-CoV-2 Spike protein in complex with a pan-sarbecovirus nanobody 2-67 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Huang, W, Taylor, D. | | 登録日 | 2022-05-23 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Superimmunity by pan-sarbecovirus nanobodies.
Cell Rep, 39, 2022
|
|
1IBR
 
 | | COMPLEX OF RAN WITH IMPORTIN BETA | | 分子名称: | GTP-binding nuclear protein RAN, Importin beta-1 subunit, MAGNESIUM ION, ... | | 著者 | Vetter, I.R, Arndt, A, Kutay, U, Goerlich, D, Wittinghofer, A. | | 登録日 | 1999-05-14 | | 公開日 | 1999-06-03 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural view of the Ran-Importin beta interaction at 2.3 A resolution
Cell(Cambridge,Mass.), 97, 1999
|
|
8CYB
 
 | |
6HL4
 
 | | wild-type NuoEF from Aquifex aeolicus - reduced form | | 分子名称: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | 著者 | Gerhardt, S, Friedrich, T, Einsle, O, Gnandt, E, Schulte, M, Fiegen, D. | | 登録日 | 2018-09-10 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
6HLI
 
 | | wild-type NuoEF from Aquifex aeolicus - reduced form bound to NAD+ | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | 著者 | Gerhardt, S, Friedrich, T, Einsle, O, Gnandt, E, Schulte, M, Fiegen, D. | | 登録日 | 2018-09-11 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
8CYJ
 
 | | RBD of SARS-CoV-2 Spike protein in complex with pan-sarbecovirus nanobodies 2-10, 2-67, 2-62 and 1-25 | | 分子名称: | Spike glycoprotein, pan-sarbecovirus nanobody 1-25, pan-sarbecovirus nanobody 2-10, ... | | 著者 | Huang, W, Taylor, D. | | 登録日 | 2022-05-23 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Superimmunity by pan-sarbecovirus nanobodies.
Cell Rep, 39, 2022
|
|
6HMC
 
 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 gene product) IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR THN27 | | 分子名称: | 1,2-ETHANEDIOL, 5-propan-2-yl-4-prop-2-enoxy-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, Casein kinase II subunit alpha' | | 著者 | Niefind, K, Lindenblatt, D, Dimper, V, Jose, J, Le Borgne, M. | | 登録日 | 2018-09-12 | | 公開日 | 2019-03-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.03 Å) | | 主引用文献 | Diacritic Binding of an Indenoindole Inhibitor by CK2 alpha Paralogs Explored by a Reliable Path to Atomic Resolution CK2 alpha ' Structures.
Acs Omega, 4, 2019
|
|
1I7R
 
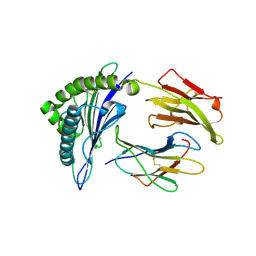 | | CRYSTAL STRUCTURE OF CLASS I MHC A2 IN COMPLEX WITH PEPTIDE P1058 | | 分子名称: | 9 RESIDUE PEPTIDE, BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | 著者 | Busslep, J, Zhao, R, Loftus, D, Appella, E, Collins, E.J. | | 登録日 | 2001-03-10 | | 公開日 | 2001-10-24 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | T cell activity correlates with oligomeric peptide/MHC binding on T cell surface
J.Biol.Chem., 276, 2001
|
|
9GA2
 
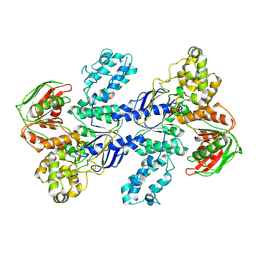 | | MtUvrA2 dimer empty | | 分子名称: | UvrABC system protein A, ZINC ION | | 著者 | Genta, M, Capelli, R, Ferrara, G, Rizzi, M, Rossi, F, Jeruzalmi, D, Bolognesi, M, Chaves-Sanjuan, A, Miggiano, R. | | 登録日 | 2024-07-26 | | 公開日 | 2025-04-23 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Mechanistic understanding of UvrA damage detection and lesion hand-off to UvrB in Nucleotide Excision Repair.
Nat Commun, 16, 2025
|
|
9GA4
 
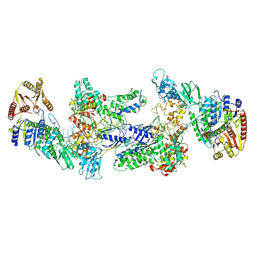 | | MtUvrA2UvrB2 bound to damaged oligonucleotide | | 分子名称: | DNA, UvrABC system protein A, UvrABC system protein B, ... | | 著者 | Genta, M, Capelli, R, Ferrara, G, Rizzi, M, Rossi, F, Jeruzalmi, D, Bolognesi, M, Chaves-Sanjuan, A, Miggiano, R. | | 登録日 | 2024-07-26 | | 公開日 | 2025-04-23 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Mechanistic understanding of UvrA damage detection and lesion hand-off to UvrB in Nucleotide Excision Repair.
Nat Commun, 16, 2025
|
|
8PK2
 
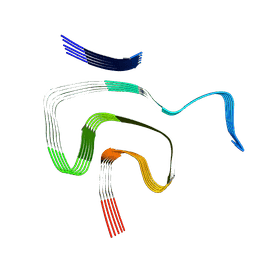 | | Cryo EM structure of the type 1m polymorph of alpha-synuclein | | 分子名称: | Alpha-synuclein | | 著者 | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | 登録日 | 2023-06-24 | | 公開日 | 2024-05-29 | | 最終更新日 | 2025-03-05 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation.
Elife, 12, 2024
|
|
5LOA
 
 | |
