8FP3
 
 | | PKCeta kinase domain in complex with compound 11 | | 分子名称: | (3P)-3-{4-[(3R,5S)-3-amino-5-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Protein kinase C eta type | | 著者 | Johnson, E. | | 登録日 | 2023-01-03 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-04-26 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FH4
 
 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undec-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile | | 分子名称: | (3P)-3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undecan-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile, Mitogen-activated protein kinase kinase kinase kinase 1, SULFATE ION | | 著者 | McTigue, M, Johnson, E, Cronin, C. | | 登録日 | 2022-12-13 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.827 Å) | | 主引用文献 | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FKO
 
 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-{4-[(2S,5R)-5-Amino-2-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile | | 分子名称: | (3P)-3-{4-[(2S,5R)-5-amino-2-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Mitogen-activated protein kinase kinase kinase kinase 1 | | 著者 | McTigue, M, Johnson, E, Cronin, C. | | 登録日 | 2022-12-21 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.104 Å) | | 主引用文献 | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FP1
 
 | | PKCeta kinase domain in complex with compound 2 | | 分子名称: | (3P)-3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undecan-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile, Protein kinase C eta type | | 著者 | Johnson, E. | | 登録日 | 2023-01-03 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-04-26 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
1GPU
 
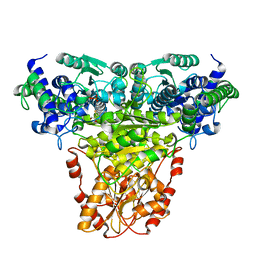 | | Transketolase complex with reaction intermediate | | 分子名称: | 2-[3-[(4-AMINO-2-METHYL-5-PYRIMIDINYL)METHYL]-2-(1,2-DIHYDROXYETHYL)-4-METHYL-1,3-THIAZOL-3-IUM-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, CALCIUM ION, TRANSKETOLASE 1 | | 著者 | Fiedler, E, Thorell, S, Sandalova, T, Koenig, S, Schneider, G. | | 登録日 | 2001-11-09 | | 公開日 | 2002-02-11 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Snapshot of a Key Intermediate in Enzymatic Thiamin Catalysis: Crystal Structure of the Alpha-Carbanion of (Alpha,Beta-Dihydroxyethyl)-Thiamin Diphosphate in the Active Site of Transketolase from Saccharomyces Cerevisiae.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
6XM5
 
 | | Structure of SARS-CoV-2 spike at pH 5.5, all RBDs down | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | 登録日 | 2020-06-29 | | 公開日 | 2020-07-29 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6XM0
 
 | | Consensus structure of SARS-CoV-2 spike at pH 5.5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-12 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6XM4
 
 | | Structure of SARS-CoV-2 spike at pH 5.5, single RBD up, conformation 2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-12 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6XLU
 
 | | Structure of SARS-CoV-2 spike at pH 4.0 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-12 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6XM3
 
 | | Structure of SARS-CoV-2 spike at pH 5.5, single RBD up, conformation 1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-12 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6URM
 
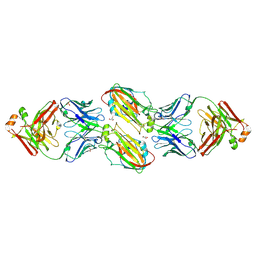 | | Crystal structure of vaccine-elicited receptor-binding site targeting antibody LPAF-a.01 in complex with Hemagglutinin H1 A/California/04/2009 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | 著者 | Zhou, T, Cheung, S.F, Kwong, P.D. | | 登録日 | 2019-10-23 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Identification and Structure of a Multidonor Class of Head-Directed Influenza-Neutralizing Antibodies Reveal the Mechanism for Its Recurrent Elicitation.
Cell Rep, 32, 2020
|
|
7R73
 
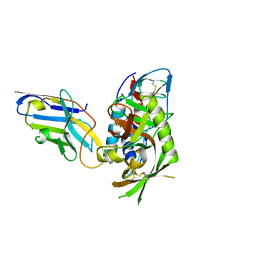 | |
7RI1
 
 | |
7RI2
 
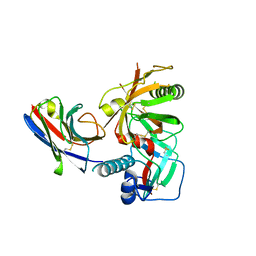 | |
7R74
 
 | |
5V7J
 
 | | Crystal Structure at 3.7 A Resolution of Glycosylated HIV-1 Clade A BG505 SOSIP.664 Prefusion Env Trimer with Four Glycans (N197, N276, N362, and N462) removed in Complex with Neutralizing Antibodies 3H+109L and 35O22. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 35O22 Fab heavy chain, ... | | 著者 | Stewart-Jones, G.B.E, Zhou, T, Kwong, P.D. | | 登録日 | 2017-03-20 | | 公開日 | 2017-06-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.907 Å) | | 主引用文献 | Quantification of the Impact of the HIV-1-Glycan Shield on Antibody Elicitation.
Cell Rep, 19, 2017
|
|
6WHC
 
 | | CryoEM Structure of the glucagon receptor with a dual-agonist peptide | | 分子名称: | Dual-agonist peptide, Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Belousoff, M.J, Sexton, P, Danev, R. | | 登録日 | 2020-04-07 | | 公開日 | 2020-05-27 | | 最終更新日 | 2020-07-22 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-electron microscopy structure of the glucagon receptor with a dual-agonist peptide.
J.Biol.Chem., 295, 2020
|
|
6NZ7
 
 | |
5F15
 
 | | Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-amino-4-deoxy-L-arabinose (L-Ara4N) transferase, CHLORIDE ION, ... | | 著者 | Petrou, V.I, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2015-11-30 | | 公開日 | 2016-02-17 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation.
Science, 351, 2016
|
|
5EZM
 
 | | Crystal Structure of ArnT from Cupriavidus metallidurans in the apo state | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferases of PMT family, CHLORIDE ION, ... | | 著者 | Petrou, V.I, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2015-11-26 | | 公開日 | 2016-02-17 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation.
Science, 351, 2016
|
|
9EYU
 
 | | Human PRMT5 in complex with AZ compound 1 | | 分子名称: | (1~{S})-2-[(2-carbamimidamido-1,3-thiazol-5-yl)methyl]-~{N}-[(4-fluorophenyl)methyl]-3-oxidanylidene-1~{H}-isoindole-1-carboxamide, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein WDR77, ... | | 著者 | Debreczeni, J. | | 登録日 | 2024-04-09 | | 公開日 | 2024-08-14 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Discovery and In Vivo Efficacy of AZ-PRMT5i-1, a Novel PRMT5 Inhibitor with High MTA Cooperativity.
J.Med.Chem., 2024
|
|
9EYV
 
 | | Human PRMT5 in complex with AZ compound 12 | | 分子名称: | (1~{S})-~{N}-[(4-fluorophenyl)methyl]-3-oxidanylidene-2-(1~{H}-pyrrolo[3,2-b]pyridin-2-ylmethyl)-1~{H}-isoindole-1-carboxamide, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein WDR77, ... | | 著者 | Debreczeni, J. | | 登録日 | 2024-04-09 | | 公開日 | 2024-08-14 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Discovery and In Vivo Efficacy of AZ-PRMT5i-1, a Novel PRMT5 Inhibitor with High MTA Cooperativity.
J.Med.Chem., 2024
|
|
9EYW
 
 | | Human PRMT5 in complex with AZ compound 21 | | 分子名称: | (3~{S})-2-[(5-azanyl-1~{H}-pyrrolo[3,2-b]pyridin-2-yl)methyl]-6-fluoranyl-1'-[(4-fluorophenyl)methyl]spiro[isoindole-3,3'-pyrrolidine]-1,2'-dione, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein WDR77, ... | | 著者 | Debreczeni, J. | | 登録日 | 2024-04-09 | | 公開日 | 2024-08-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Discovery and In Vivo Efficacy of AZ-PRMT5i-1, a Novel PRMT5 Inhibitor with High MTA Cooperativity.
J.Med.Chem., 2024
|
|
9EYX
 
 | | Human PRMT5 in complex with AZ compound 28 | | 分子名称: | (3~{S})-2-[(5-azanyl-6-fluoranyl-1~{H}-pyrrolo[3,2-b]pyridin-2-yl)methyl]-6-fluoranyl-1'-[(4-fluorophenyl)methyl]spiro[isoindole-3,3'-pyrrolidine]-1,2'-dione, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein WDR77, ... | | 著者 | Debreczeni, J. | | 登録日 | 2024-04-09 | | 公開日 | 2024-08-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Discovery and In Vivo Efficacy of AZ-PRMT5i-1, a Novel PRMT5 Inhibitor with High MTA Cooperativity.
J.Med.Chem., 2024
|
|
4YQH
 
 | | 2-[2-(4-Phenyl-1H-imidazol-2-yl)ethyl]quinoxaline (Sunovion Compound 14) co-crystallized with PDE10A | | 分子名称: | 2-[2-(4-phenyl-1H-imidazol-2-yl)ethyl]quinoxaline, MAGNESIUM ION, ZINC ION, ... | | 著者 | Burdi, D, Herman, L, Wang, T. | | 登録日 | 2015-03-13 | | 公開日 | 2015-04-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.308 Å) | | 主引用文献 | Evolution and synthesis of novel orally bioavailable inhibitors of PDE10A.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
