8J21
 
 | | Cryo-EM structure of FFAR3 complex bound with butyrate acid | | 分子名称: | Free fatty acid receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | 登録日 | 2023-04-14 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
7XLB
 
 | |
6JX1
 
 | | Crystal structure of Formate dehydrogenase mutant V198I/C256I/P260S/E261P/S381N/S383F from Pseudomonas sp. 101 | | 分子名称: | Formate dehydrogenase, GLYCEROL | | 著者 | Feng, Y, Xue, S, Guo, X, Zhao, Z. | | 登録日 | 2019-04-21 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.233 Å) | | 主引用文献 | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
6JUK
 
 | | Crystal structure of Formate dehydrogenase mutant C256I/E261P/S381I from Pseudomonas sp. 101 in complex with non-natural cofactor Nicotinamide Cytosine Dinucleotide | | 分子名称: | Formate dehydrogenase, GLYCEROL, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Feng, Y, Xue, S, Guo, X, Zhao, Z. | | 登録日 | 2019-04-14 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.293 Å) | | 主引用文献 | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
6JUJ
 
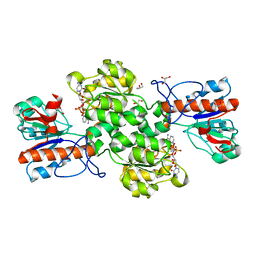 | | Crystal structure of Formate dehydrogenase mutant V198I/C256I/P260S/E261P/S381N/S383F from Pseudomonas sp. 101in complex with non-natural cofactor Nicotinamide Cytosine Dinucleotide | | 分子名称: | Formate dehydrogenase, GLYCEROL, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Feng, Y, Guo, X, Xue, S, Zhao, Z. | | 登録日 | 2019-04-14 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.183 Å) | | 主引用文献 | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
6JWG
 
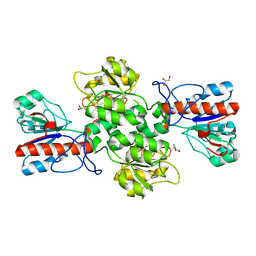 | | Crystal structure of Formate dehydrogenase mutant C256I/E261P/S381I from Pseudomonas sp. 101 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Formate dehydrogenase, GLYCEROL | | 著者 | Feng, Y, Guo, X, Xue, S, Zhao, Z. | | 登録日 | 2019-04-20 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.081 Å) | | 主引用文献 | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
2GD3
 
 | | NMR structure of S14G-humanin in 30% TFE solution | | 分子名称: | Humanin | | 著者 | Benaki, D, Zikos, C, Evangelou, A, Livaniou, E, Vlassi, M, Mikros, E, Pelecanou, M. | | 登録日 | 2006-03-15 | | 公開日 | 2006-09-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of Ser14Gly-humanin, a potent rescue factor against neuronal cell death in Alzheimer's disease.
Biochem.Biophys.Res.Commun., 349, 2006
|
|
7D8T
 
 | | MITF bHLHLZ complex with M-box DNA | | 分子名称: | DNA (5'-D(*TP*GP*TP*AP*AP*CP*AP*TP*GP*TP*GP*TP*CP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*AP*CP*AP*CP*AP*TP*GP*TP*TP*AP*CP*AP*G)-3'), Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit | | 著者 | Guo, M, Fang, P, Wang, J. | | 登録日 | 2020-10-09 | | 公開日 | 2021-10-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.201 Å) | | 主引用文献 | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7D8R
 
 | | MITF HLHLZ structure | | 分子名称: | Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit | | 著者 | Guo, M, Fang, P, Wang, J. | | 登録日 | 2020-10-09 | | 公開日 | 2021-10-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7D8S
 
 | | MITF bHLHLZ apo structure | | 分子名称: | Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit, SULFATE ION | | 著者 | Guo, M, Fang, P, Wang, J. | | 登録日 | 2020-10-09 | | 公開日 | 2021-10-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7BW6
 
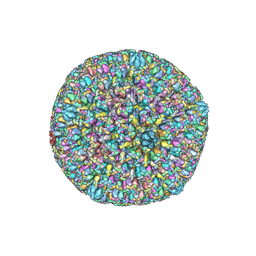 | | Varicella-zoster virus capsid | | 分子名称: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | 著者 | Wang, P.Y, Qi, J.X, Liu, C.C, Sun, J.Q. | | 登録日 | 2020-04-13 | | 公開日 | 2020-09-23 | | 最終更新日 | 2020-10-07 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structure of the varicella-zoster virus A-capsid.
Nat Commun, 11, 2020
|
|
8JB5
 
 | |
7X2D
 
 | | Cryo-EM structure of the tavapadon-bound D1 dopamine receptor and mini-Gs complex | | 分子名称: | 1,5-dimethyl-6-[2-methyl-4-[3-(trifluoromethyl)pyridin-2-yl]oxy-phenyl]pyrimidine-2,4-dione, CHOLESTEROL, D(1A) dopamine receptor, ... | | 著者 | Teng, X, Zheng, S. | | 登録日 | 2022-02-25 | | 公開日 | 2022-06-15 | | 最終更新日 | 2022-10-12 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Ligand recognition and biased agonism of the D1 dopamine receptor.
Nat Commun, 13, 2022
|
|
7X2F
 
 | | Cryo-EM structure of the dopamine and LY3154207-bound D1 dopamine receptor and mini-Gs complex | | 分子名称: | 2-[2,6-bis(chloranyl)phenyl]-1-[(1S,3R)-3-(hydroxymethyl)-1-methyl-5-(3-methyl-3-oxidanyl-butyl)-3,4-dihydro-1H-isoquinolin-2-yl]ethanone, CHOLESTEROL, D(1A) dopamine receptor, ... | | 著者 | Teng, X, Zheng, S. | | 登録日 | 2022-02-25 | | 公開日 | 2022-06-15 | | 最終更新日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Ligand recognition and biased agonism of the D1 dopamine receptor.
Nat Commun, 13, 2022
|
|
7X2C
 
 | | Cryo-EM structure of the fenoldopam-bound D1 dopamine receptor and mini-Gs complex | | 分子名称: | (1R)-6-chloranyl-1-(4-hydroxyphenyl)-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | 著者 | Teng, X, Zheng, S. | | 登録日 | 2022-02-25 | | 公開日 | 2022-06-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Ligand recognition and biased agonism of the D1 dopamine receptor.
Nat Commun, 13, 2022
|
|
7WA9
 
 | |
6KHY
 
 | | The crystal structure of AsfvAP:AG | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (AGCGTCACCGACGAGGC), DNA(AGCGTCACCGACGAGG), ... | | 著者 | Chen, Y.Q, Gan, J.H. | | 登録日 | 2019-07-16 | | 公開日 | 2020-06-03 | | 実験手法 | X-RAY DIFFRACTION (3.008 Å) | | 主引用文献 | A unique DNA-binding mode of African swine fever virus AP endonuclease.
Cell Discov, 6, 2020
|
|
6KI3
 
 | | The crystal structure of AsfvAP:dF commplex | | 分子名称: | DNA (5'-D(*CP*CP*TP*CP*GP*TP*CP*GP*GP*GP*GP*AP*CP*GP*CP*TP*G)-3'), DNA (5'-D(*GP*CP*AP*GP*CP*GP*TP*CP*C)-3'), DNA (5'-D(P*(3DR)P*CP*GP*AP*CP*GP*AP*G)-3'), ... | | 著者 | Chen, Y, Gan, J. | | 登録日 | 2019-07-17 | | 公開日 | 2020-05-27 | | 実験手法 | X-RAY DIFFRACTION (2.354 Å) | | 主引用文献 | A unique DNA-binding mode of African swine fever virus AP endonuclease.
Cell Discov, 6, 2020
|
|
8J1F
 
 | | GSK101 bound state of mTRPV4 | | 分子名称: | N-[(2S)-1-{4-[N-(2,4-dichlorobenzene-1-sulfonyl)-L-seryl]piperazin-1-yl}-4-methyl-1-oxopentan-2-yl]-1-benzothiophene-2-carboxamide, Transient receptor potential cation channel subfamily V member 4 | | 著者 | Zhen, W.X, Yang, F. | | 登録日 | 2023-04-12 | | 公開日 | 2023-08-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.62 Å) | | 主引用文献 | Structural basis of ligand activation and inhibition in a mammalian TRPV4 ion channel.
Cell Discov, 9, 2023
|
|
8J1H
 
 | | Agonist1 and Ruthenium Red bound state of mTRPV4 | | 分子名称: | 8-fluoranyl-3-[4-(4-fluoranylphenoxy)phenyl]-2-(4-methylpiperazin-1-yl)quinazolin-4-one, Transient receptor potential cation channel subfamily V member 4 | | 著者 | Zhen, W.X, Yang, F. | | 登録日 | 2023-04-12 | | 公開日 | 2023-08-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.88 Å) | | 主引用文献 | Structural basis of ligand activation and inhibition in a mammalian TRPV4 ion channel.
Cell Discov, 9, 2023
|
|
8J1D
 
 | | Cryo-EM structure of apo state mTRPV4 | | 分子名称: | Transient receptor potential cation channel subfamily V member 4 | | 著者 | Zhen, W.X, Yang, F. | | 登録日 | 2023-04-12 | | 公開日 | 2023-08-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.59 Å) | | 主引用文献 | Structural basis of ligand activation and inhibition in a mammalian TRPV4 ion channel.
Cell Discov, 9, 2023
|
|
8J1B
 
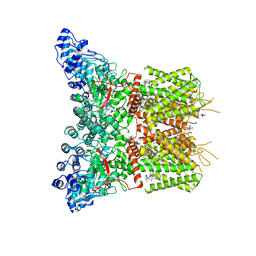 | | GSK101 and Ruthenium Red bound state of mTRPV4 | | 分子名称: | N-[(2S)-1-{4-[N-(2,4-dichlorobenzene-1-sulfonyl)-L-seryl]piperazin-1-yl}-4-methyl-1-oxopentan-2-yl]-1-benzothiophene-2-carboxamide, Transient receptor potential cation channel subfamily V member 4, ruthenium(6+) azanide pentaamino(oxido)ruthenium (1/4/2) | | 著者 | Zhen, W.X, Yang, F. | | 登録日 | 2023-04-12 | | 公開日 | 2023-08-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.72 Å) | | 主引用文献 | Structural basis of ligand activation and inhibition in a mammalian TRPV4 ion channel.
Cell Discov, 9, 2023
|
|
8KA5
 
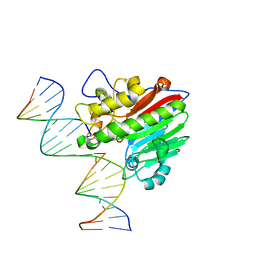 | |
8KA3
 
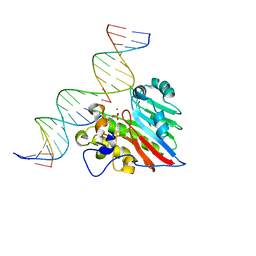 | |
8KA4
 
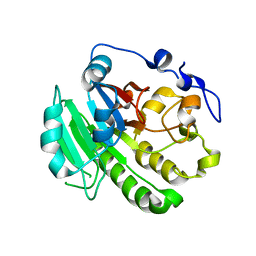 | |
