8Z8S
 
 | |
8Z8P
 
 | |
9BR6
 
 | |
9BR7
 
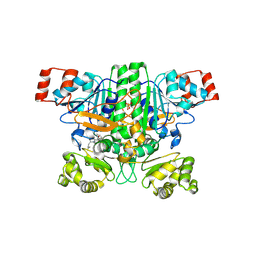 | | Crystal structure of human succinyl-CoA:glutarate-CoA transferase (SUGCT) in complex with Losartan carboxylic acid | | 分子名称: | AMMONIUM ION, SULFATE ION, Succinate--hydroxymethylglutarate CoA-transferase, ... | | 著者 | Wu, R, Lazarus, M.B. | | 登録日 | 2024-05-10 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Characterization, Structure, and Inhibition of the Human Succinyl-CoA:glutarate-CoA Transferase, a Putative Genetic Modifier of Glutaric Aciduria Type 1.
Acs Chem.Biol., 19, 2024
|
|
8Z8R
 
 | |
8Z8O
 
 | |
3FKR
 
 | |
3FKK
 
 | | Structure of L-2-keto-3-deoxyarabonate dehydratase | | 分子名称: | L-2-keto-3-deoxyarabonate dehydratase, PHOSPHATE ION | | 著者 | Shimada, N, Mikami, B. | | 登録日 | 2008-12-17 | | 公開日 | 2010-01-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural analysis of L -2-keto-3-deoxyarabonate dehydratase an enzyme involved in an alternative bacterial pathway of L-arabinose metabolism in complex with pyruvate
To be Published
|
|
3IQI
 
 | |
4LDD
 
 | |
4LDM
 
 | |
4LDI
 
 | |
4LDB
 
 | |
4LD8
 
 | |
3IQG
 
 | |
1YNR
 
 | | Crystal structure of the cytochrome c-552 from Hydrogenobacter thermophilus at 2.0 resolution | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cytochrome c-552, HEME C, ... | | 著者 | Travaglini-Allocatelli, C, Gianni, S, Dubey, V.K, Borgia, A, Di Matteo, A, Bonivento, D, Cutruzzola, F, Bren, K.L, Brunori, M. | | 登録日 | 2005-01-25 | | 公開日 | 2005-05-17 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | An Obligatory Intermediate in the Folding Pathway of Cytochrome c552 from Hydrogenobacter thermophilus
J.Biol.Chem., 280, 2005
|
|
3IQH
 
 | |
1QNE
 
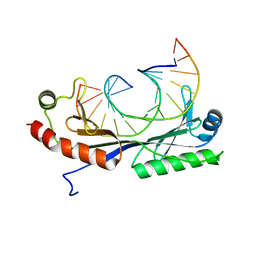 | |
6AI6
 
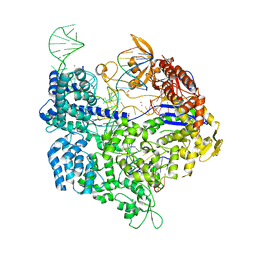 | | Crystal structure of SpCas9-NG | | 分子名称: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9/Csn1, DNA (28-MER), ... | | 著者 | Nishimasu, H, Hirano, S, Ishitani, R, Nureki, O. | | 登録日 | 2018-08-21 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Engineered CRISPR-Cas9 nuclease with expanded targeting space
Science, 361, 2018
|
|
7W6F
 
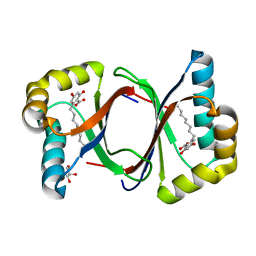 | | Polyketide cyclase OAC-F24I mutant from Cannabis sativa in complex with 6-nonylresorcylic acid | | 分子名称: | 2-nonyl-4,6-bis(oxidanyl)benzoic acid, GLYCEROL, Olivetolic acid cyclase | | 著者 | Nakashima, Y, Lee, Y.E, Morita, H. | | 登録日 | 2021-12-01 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Dual Engineering of Olivetolic Acid Cyclase and Tetraketide Synthase to Generate Longer Alkyl-Chain Olivetolic Acid Analogs.
Org.Lett., 24, 2022
|
|
7W6G
 
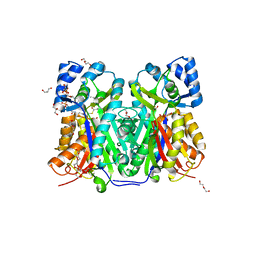 | | TKS-L190G mutant from Cannabis sativa in complex with lauroyl-CoA | | 分子名称: | 3,5,7-trioxododecanoyl-CoA synthase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Nakashima, Y, Lee, Y.E, Morita, H. | | 登録日 | 2021-12-01 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Dual Engineering of Olivetolic Acid Cyclase and Tetraketide Synthase to Generate Longer Alkyl-Chain Olivetolic Acid Analogs.
Org.Lett., 24, 2022
|
|
7W6E
 
 | |
7W6D
 
 | |
7Y3V
 
 | | Crystal structure of CdpNPT in complex with harmane | | 分子名称: | 1-methyl-9H-pyrido[3,4-b]indole, Cyclic dipeptide N-prenyltransferase, PHOSPHATE ION | | 著者 | Nakashima, Y, Morita, H. | | 登録日 | 2022-06-13 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Catalytic potential of a fungal indole prenyltransferase toward beta-carbolines, harmine and harman, and their prenylation effects on antibacterial activity.
J.Biosci.Bioeng., 134, 2022
|
|
7EJL
 
 | |
