8I2R
 
 | |
8KBE
 
 | | Structure of CbTad1 complexed with 1',3'-cADPR | | 分子名称: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, Thoeris anti-defense 1 | | 著者 | Xiao, Y, Feng, Y. | | 登録日 | 2023-08-04 | | 公開日 | 2024-08-07 | | 最終更新日 | 2024-12-04 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Single phage proteins sequester signals from TIR and cGAS-like enzymes.
Nature, 635, 2024
|
|
8KBF
 
 | | Structure of CbTad1 complexed with 1',3'-cADPR and cA3 | | 分子名称: | (2-ACETYL-5-METHYLANILINO)(2,6-DIBROMOPHENYL)ACETAMIDE, (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, Thoeris anti-defense 1 | | 著者 | Xiao, Y, Feng, Y. | | 登録日 | 2023-08-04 | | 公開日 | 2024-08-07 | | 最終更新日 | 2024-12-04 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Single phage proteins sequester signals from TIR and cGAS-like enzymes.
Nature, 635, 2024
|
|
8KBB
 
 | | Structure of apo-CmTad1 | | 分子名称: | Thoeris anti-defense 1, ZINC ION | | 著者 | Xiao, Y, Feng, Y. | | 登録日 | 2023-08-04 | | 公開日 | 2024-08-07 | | 最終更新日 | 2024-12-04 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Single phage proteins sequester signals from TIR and cGAS-like enzymes.
Nature, 635, 2024
|
|
8KBL
 
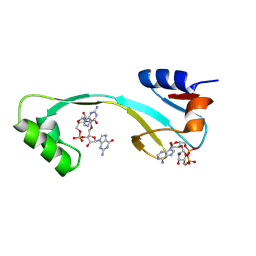 | | Structure of AcrIIA7 complexed with 1',3'-cADPR and cGG | | 分子名称: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Inhibitor of Type II CRISPR-Cas system | | 著者 | Xiao, Y, Feng, Y. | | 登録日 | 2023-08-04 | | 公開日 | 2024-08-07 | | 最終更新日 | 2024-12-04 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Single phage proteins sequester signals from TIR and cGAS-like enzymes.
Nature, 635, 2024
|
|
8KBK
 
 | | Structure of AcrIIA7 complexed with 1',2'-cADPR and cGG | | 分子名称: | (1S,3R,4R,6R,9S,11R,14R,15S,16R,18R)-4-(6-amino-9H-purin-9-yl)-9,11,15,16,18-pentahydroxy-2,5,8,10,12,17-hexaoxa-9lambda~5~,11lambda~5~-diphosphatricyclo[12.2.1.1~3,6~]octadecane-9,11-dione, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Inhibitor of Type II CRISPR-Cas system | | 著者 | Xiao, Y, Feng, Y. | | 登録日 | 2023-08-04 | | 公開日 | 2024-08-07 | | 最終更新日 | 2024-12-04 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Single phage proteins sequester signals from TIR and cGAS-like enzymes.
Nature, 635, 2024
|
|
8KBD
 
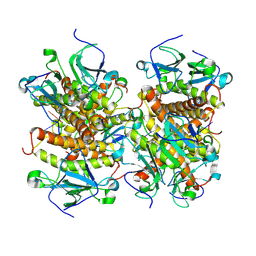 | |
8KBI
 
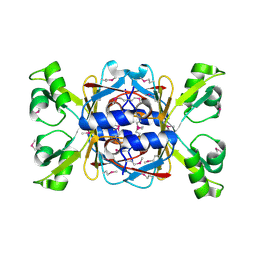 | | Structure of apo-AcrIIA7 | | 分子名称: | Inhibitor of Type II CRISPR-Cas system | | 著者 | Xiao, Y, Feng, Y. | | 登録日 | 2023-08-04 | | 公開日 | 2024-08-07 | | 最終更新日 | 2024-12-04 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Single phage proteins sequester signals from TIR and cGAS-like enzymes.
Nature, 635, 2024
|
|
8KBM
 
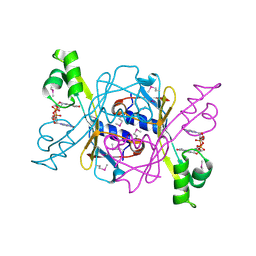 | | Structure of AcrIIA7 complexed with cGG | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Inhibitor of Type II CRISPR-Cas system | | 著者 | Xiao, Y, Feng, Y. | | 登録日 | 2023-08-04 | | 公開日 | 2024-08-07 | | 最終更新日 | 2024-12-04 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Single phage proteins sequester signals from TIR and cGAS-like enzymes.
Nature, 635, 2024
|
|
8KBJ
 
 | | Structure of HgmTad2 complexed with 1',2'-cADPR | | 分子名称: | (1S,3R,4R,6R,9S,11R,14R,15S,16R,18R)-4-(6-amino-9H-purin-9-yl)-9,11,15,16,18-pentahydroxy-2,5,8,10,12,17-hexaoxa-9lambda~5~,11lambda~5~-diphosphatricyclo[12.2.1.1~3,6~]octadecane-9,11-dione, Inhibitor of Type II CRISPR-Cas system | | 著者 | Xiao, Y, Feng, Y. | | 登録日 | 2023-08-04 | | 公開日 | 2024-08-21 | | 最終更新日 | 2024-12-04 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Single phage proteins sequester signals from TIR and cGAS-like enzymes.
Nature, 635, 2024
|
|
8KBG
 
 | |
8KBC
 
 | | Structure of CmTad1 complexed with cAAA | | 分子名称: | (2-ACETYL-5-METHYLANILINO)(2,6-DIBROMOPHENYL)ACETAMIDE, Thoeris anti-defense 1, ZINC ION | | 著者 | Xiao, Y, Feng, Y. | | 登録日 | 2023-08-04 | | 公開日 | 2024-08-21 | | 最終更新日 | 2024-12-04 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Single phage proteins sequester signals from TIR and cGAS-like enzymes.
Nature, 635, 2024
|
|
8KBH
 
 | |
8WJE
 
 | | Structure of apo-SPO1 Tad2 | | 分子名称: | Gp34.65 | | 著者 | Xiao, Y, Feng, Y. | | 登録日 | 2023-09-25 | | 公開日 | 2024-09-25 | | 最終更新日 | 2024-12-04 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Single phage proteins sequester signals from TIR and cGAS-like enzymes.
Nature, 635, 2024
|
|
8WJD
 
 | | Structure of SptTad2 complexed with cGG | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), SptTad2 | | 著者 | Xiao, Y, Feng, Y. | | 登録日 | 2023-09-25 | | 公開日 | 2024-09-25 | | 最終更新日 | 2024-12-04 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Single phage proteins sequester signals from TIR and cGAS-like enzymes.
Nature, 635, 2024
|
|
8WJC
 
 | | Structure of AcrIIA7 complexed with 3'3'-cGAMP | | 分子名称: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Inhibitor of Type II CRISPR-Cas system | | 著者 | Xiao, Y, Feng, Y. | | 登録日 | 2023-09-25 | | 公開日 | 2024-09-25 | | 最終更新日 | 2024-12-04 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Single phage proteins sequester signals from TIR and cGAS-like enzymes.
Nature, 635, 2024
|
|
7C8P
 
 | |
9B4E
 
 | | Structure of wild type human PSS1 | | 分子名称: | (2R)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dihexadecanoate, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CALCIUM ION, ... | | 著者 | Long, T, Li, X. | | 登録日 | 2024-03-20 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Molecular insights into human phosphatidylserine synthase 1 reveal its inhibition promotes LDL uptake.
Cell, 187, 2024
|
|
9B4G
 
 | | Structure of inhibitor-bound human PSS1 | | 分子名称: | (2R)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dihexadecanoate, (7P)-7-[(4S)-4-{4-[3,5-bis(trifluoromethyl)phenoxy]phenyl}-5-(2,2-difluoropropyl)-6-oxo-1,4,5,6-tetrahydropyrrolo[3,4-c]pyrazol-3-yl]-1,3-benzoxazol-2(3H)-one, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, ... | | 著者 | Long, T, Li, X. | | 登録日 | 2024-03-20 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (2.87 Å) | | 主引用文献 | Molecular insights into human phosphatidylserine synthase 1 reveal its inhibition promotes LDL uptake.
Cell, 187, 2024
|
|
9B4F
 
 | | Structure of human PSS1-P269S | | 分子名称: | CALCIUM ION, Phosphatidylserine synthase 1 | | 著者 | Long, T, Li, X. | | 登録日 | 2024-03-20 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | Molecular insights into human phosphatidylserine synthase 1 reveal its inhibition promotes LDL uptake.
Cell, 187, 2024
|
|
9JL0
 
 | | Crystal structure of feline CD8aa homodimer | | 分子名称: | T-cell surface glycoprotein CD8 alpha chain | | 著者 | Li, Z, Liang, R. | | 登録日 | 2024-09-17 | | 公開日 | 2025-07-02 | | 最終更新日 | 2025-07-09 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Insights into the structural features of a feline CD8 alpha alpha homodimer.
Dev.Comp.Immunol., 2025
|
|
8HRD
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Delta variant in complex with IMCAS74 Fab and W14 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS74 Fab heavy chain, IMCAS74 Fab light chain, ... | | 著者 | Zhao, R.C, Wu, L.L, Han, P. | | 登録日 | 2022-12-15 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.86 Å) | | 主引用文献 | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6KXW
 
 | | Crystal structure of human aquaporin AQP7 in bound to glycerol | | 分子名称: | Aquaporin-7, GLYCEROL | | 著者 | Zhang, L, Yao, D, Zhou, F, Zhang, Q, Zhou, L, Cao, Y. | | 登録日 | 2019-09-13 | | 公開日 | 2019-11-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | The structural basis for glycerol permeation by human AQP7
Sci Bull (Beijing), 66, 2020
|
|
4WBX
 
 | | Conserved hypothetical protein PF1771 from Pyrococcus furiosus solved by sulfur SAD using Swiss Light Source data | | 分子名称: | 2-keto acid:ferredoxin oxidoreductase subunit alpha | | 著者 | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2014-09-04 | | 公開日 | 2014-12-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.301 Å) | | 主引用文献 | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
5X5Y
 
 | | A membrane protein complex | | 分子名称: | Probable ATP-binding component of ABC transporter, Uncharacterized protein | | 著者 | Luo, Q, Yang, X, Huang, Y. | | 登録日 | 2017-02-18 | | 公開日 | 2017-04-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.465 Å) | | 主引用文献 | Structural basis for lipopolysaccharide extraction by ABC transporter LptB2FG
Nat. Struct. Mol. Biol., 24, 2017
|
|
