6X5P
 
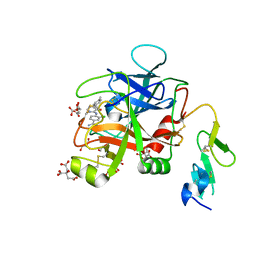 | | Discovery of Hydroxy Pyrimidine Factor IXa Inhibitors | | 分子名称: | 3-chloro-4-{[5-hydroxy-6-(4-methylphenyl)pyrimidin-4-yl]amino}benzene-1-carboximidamide, CITRIC ACID, Coagulation factor IX, ... | | 著者 | Jayne, C.L, Andreani, T, Chan, T, Chelliah, M.V, Clasby, M.C, Dwyer, M, Eagen, K.A, Fried, S, Greenlee, W.J, Guo, Z, Hawes, B, Hruza, A, Ingram, R, Keertikar, K.M, Neelamkavil, S, Reichert, P, Xia, Y, Chackalamannil, S. | | 登録日 | 2020-05-26 | | 公開日 | 2020-06-24 | | 実験手法 | X-RAY DIFFRACTION (1.997 Å) | | 主引用文献 | Discovery of hydroxy pyrimidine Factor IXa inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6GF1
 
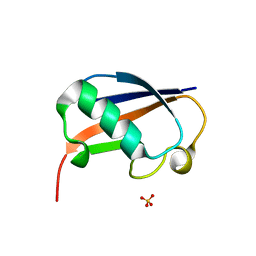 | | The structure of the ubiquitin-like modifier FAT10 reveals a novel targeting mechanism for degradation by the 26S proteasome | | 分子名称: | SULFATE ION, Ubiquitin D | | 著者 | Aichem, A, Anders, S, Catone, N, Roessler, P, Stotz, S, Berg, A, Schwab, R, Scheuermann, S, Bialas, J, Schmidtke, G, Peter, C, Groettrup, M, Wiesner, S. | | 登録日 | 2018-04-28 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.925 Å) | | 主引用文献 | The structure of the ubiquitin-like modifier FAT10 reveals an alternative targeting mechanism for proteasomal degradation.
Nat Commun, 9, 2018
|
|
4GEQ
 
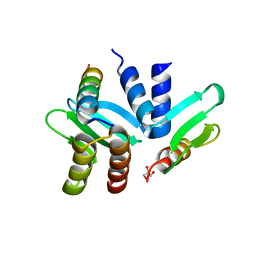 | | Crystal structure of the Spc24-Spc25/Cnn1 binding interface | | 分子名称: | GLYCEROL, Kinetochore protein SPC24, Kinetochore protein SPC25, ... | | 著者 | Malvezzi, F, Litos, G, Schleiffer, A, Heuck, A, Clausen, T, Westermann, S. | | 登録日 | 2012-08-02 | | 公開日 | 2013-01-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | A structural basis for kinetochore recruitment of the Ndc80 complex via two distinct centromere receptors.
Embo J., 32, 2013
|
|
4I9Y
 
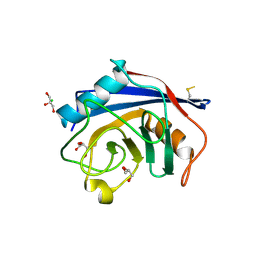 | | Structure of the C-terminal domain of Nup358 | | 分子名称: | CHLORIDE ION, E3 SUMO-protein ligase RanBP2, GLYCEROL, ... | | 著者 | Lin, D.H, Zimmermann, S, Stuwe, T, Stuwe, E, Hoelz, A. | | 登録日 | 2012-12-05 | | 公開日 | 2013-01-30 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural and Functional Analysis of the C-Terminal Domain of Nup358/RanBP2.
J.Mol.Biol., 425, 2013
|
|
8A5Q
 
 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of Chaetomium thermophilum INO80 on straight DNA | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin related protein 4 (Arp4), ... | | 著者 | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | 登録日 | 2022-06-15 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
8A5A
 
 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of INO80 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-like protein ARP8, ... | | 著者 | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | 登録日 | 2022-06-14 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
8A5P
 
 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of Chaetomium thermophilum INO80 on curved DNA | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin related protein 4 (Arp4), ... | | 著者 | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | 登録日 | 2022-06-15 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
8A5D
 
 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of Chaetomium thermophilum INO80 | | 分子名称: | Actin, Actin related protein 4 (Arp4), Actin-related protein 8, ... | | 著者 | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | 登録日 | 2022-06-14 | | 公開日 | 2022-12-28 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
8Q3J
 
 | | Crystal structure of mIL-38 in complex with a neutralizing Fab e04 fragment | | 分子名称: | Fab e04 Heavy Chain (e04 HC), Fab e04 Light Chain (e04 LC), Interleukin-1 family member 10, ... | | 著者 | Garcia-Pardo, J, Da Silva, P, Mora, J, Wiechmann, S, Putyrski, M, You, X, Kannt, A, Ernst, A, Brune, B, Weigert, A. | | 登録日 | 2023-08-04 | | 公開日 | 2024-08-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Neutralizing IL-38 activates gammaDelta T cell-dependent anti-tumor immunity and sensitizes for chemotherapy
To be published
|
|
8ATF
 
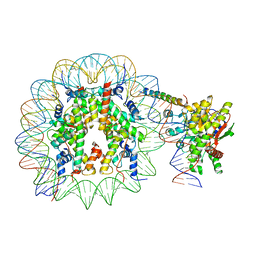 | | Nucleosome-bound Ino80 ATPase | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (226-MER), DNA (227-MER), ... | | 著者 | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | 登録日 | 2022-08-23 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.45 Å) | | 主引用文献 | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
8AV6
 
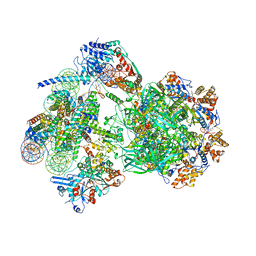 | | CryoEM structure of INO80 core nucleosome complex in closed grappler conformation | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DASH complex subunit DAD4, ... | | 著者 | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | 登録日 | 2022-08-26 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.68 Å) | | 主引用文献 | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
6E88
 
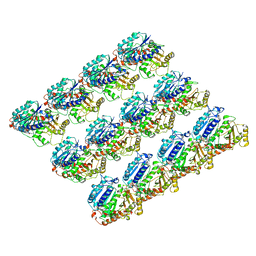 | | Cryo-EM structure of C. elegans GDP-microtubule | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Tubulin alpha-2 chain, ... | | 著者 | Chaaban, S, Jariwala, S, Chieh-Ting, H, Redemann, S, Kollman, J, Muller-Reichert, T, Sept, D, Bui, K.H, Brouhard, G.J. | | 登録日 | 2018-07-27 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | The Structure and Dynamics of C. elegans Tubulin Reveals the Mechanistic Basis of Microtubule Growth.
Dev. Cell, 47, 2018
|
|
2UY8
 
 | | R92A mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | 著者 | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | 登録日 | 2007-04-03 | | 公開日 | 2007-08-21 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
2UY9
 
 | | E162A mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | 分子名称: | MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | 著者 | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | 登録日 | 2007-04-03 | | 公開日 | 2007-08-21 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
2V09
 
 | | SENS161-164DSSN mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | 著者 | Burrell, M.R, Just, V.J, Bowater, L, Fairhurst, S.A, Requena, L, Lawson, D.M, Bornemann, S. | | 登録日 | 2007-05-10 | | 公開日 | 2007-10-30 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Oxalate Decarboxylase and Oxalate Oxidase Activities Can be Interchanged with a Specificity Switch of Up to 282 000 by Mutating an Active Site Lid.
Biochemistry, 46, 2007
|
|
2W2E
 
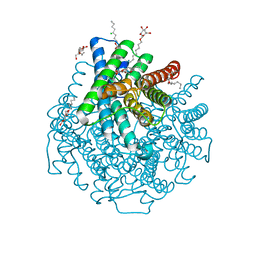 | | 1.15 Angstrom crystal structure of P.pastoris aquaporin, Aqy1, in a closed conformation at pH 3.5 | | 分子名称: | AQUAPORIN PIP2-7 7, CHLORIDE ION, octyl beta-D-glucopyranoside | | 著者 | Fischer, G, Kosinska-Eriksson, U, Aponte-Santamaria, C, Palmgren, M, Geijer, C, Hedfalk, K, Hohmann, S, de Groot, B.L, Neutze, R, Lindkvist-Petersson, K. | | 登録日 | 2008-10-29 | | 公開日 | 2009-06-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Crystal Structure of a Yeast Aquaporin at 1.15 A Reveals a Novel Gating Mechanism.1.15 A
Plos Biol., 7, 2009
|
|
2UYA
 
 | | DEL162-163 mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | 登録日 | 2007-04-03 | | 公開日 | 2007-08-21 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
2W1P
 
 | | 1.4 Angstrom crystal structure of P.pastoris aquaporin, Aqy1, in a closed conformation at pH 8.0 | | 分子名称: | AQUAPORIN PIP2-7 7;, CHLORIDE ION, octyl beta-D-glucopyranoside | | 著者 | Fischer, G, Kosinska-Eriksson, U, Aponte-Santamaria, C, Palmgren, M, Geijer, C, Hedfalk, K, Hohmann, S, de Groot, B.L, Neutze, R, Lindkvist-Petersson, K. | | 登録日 | 2008-10-20 | | 公開日 | 2009-06-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal Structure of a Yeast Aquaporin at 1.15 A Reveals a Novel Gating Mechanism
Plos Biol., 7, 2009
|
|
6FKR
 
 | | Crystal structure of the dolphin proline-rich antimicrobial peptide Tur1A bound to the Thermus thermophilus 70S ribosome | | 分子名称: | 16 ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Mardirossian, M, Perebaskine, N, Benincasa, M, Gambato, S, Hofmann, S, Huter, P, Muller, C, Hilpert, K, Innis, C.A, Tossi, A, Wilson, D.N. | | 登録日 | 2018-01-24 | | 公開日 | 2018-03-28 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | The Dolphin Proline-Rich Antimicrobial Peptide Tur1A Inhibits Protein Synthesis by Targeting the Bacterial Ribosome.
Cell Chem Biol, 25, 2018
|
|
2UYB
 
 | | S161A mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, MANGANESE (II) ION, ... | | 著者 | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | 登録日 | 2007-04-03 | | 公開日 | 2007-08-21 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
4A0O
 
 | | Symmetry-free cryo-EM map of TRiC in the nucleotide-free (apo) state | | 分子名称: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | 著者 | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | 登録日 | 2011-09-10 | | 公開日 | 2012-02-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (10.5 Å) | | 主引用文献 | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
4A0W
 
 | | model built against symmetry-free cryo-EM map of TRiC-ADP-AlFx | | 分子名称: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | 著者 | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | 登録日 | 2011-09-13 | | 公開日 | 2012-02-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (13.9 Å) | | 主引用文献 | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
4A13
 
 | | model refined against symmetry-free cryo-EM map of TRiC-ADP | | 分子名称: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | 著者 | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | 登録日 | 2011-09-13 | | 公開日 | 2012-02-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (11.3 Å) | | 主引用文献 | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
4A0V
 
 | | model refined against the Symmetry-free cryo-EM map of TRiC-AMP-PNP | | 分子名称: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | 著者 | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | 登録日 | 2011-09-13 | | 公開日 | 2012-02-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (10.7 Å) | | 主引用文献 | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
8CLJ
 
 | | TFIIIC TauB-DNA dimer | | 分子名称: | General transcription factor 3C polypeptide 1, General transcription factor 3C polypeptide 2, General transcription factor 3C polypeptide 4, ... | | 著者 | Seifert-Davila, W, Girbig, M, Hauptmann, L, Hoffmann, T, Eustermann, S, Mueller, C.W. | | 登録日 | 2023-02-16 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural insights into human TFIIIC promoter recognition.
Sci Adv, 9, 2023
|
|
