7OUY
 
 | | Crystal structure of dimeric chlorite dismutase variant R127A (CCld R127A) from Cyanothece sp. PCC7425 in complex with nitrite | | 分子名称: | Chlorite dismutase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | 登録日 | 2021-06-14 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Impact of the dynamics of the catalytic arginine on nitrite and chlorite binding by dimeric chlorite dismutase.
J.Inorg.Biochem., 227, 2021
|
|
7ATI
 
 | | Crystal structure of dimeric chlorite dismutase variant Q74V (CCld Q74V) from Cyanothece sp. PCC7425 | | 分子名称: | Chlorite dismutase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | 登録日 | 2020-10-30 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Arresting the Catalytic Arginine in Chlorite Dismutases: Impact on Heme Coordination, Thermal Stability, and Catalysis.
Biochemistry, 60, 2021
|
|
7ASB
 
 | | Crystal structure of dimeric chlorite dismutase variant Q74E (CCld Q74E) from Cyanothece sp. PCC7425 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chlorite dismutase, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | 登録日 | 2020-10-27 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Arresting the Catalytic Arginine in Chlorite Dismutases: Impact on Heme Coordination, Thermal Stability, and Catalysis.
Biochemistry, 60, 2021
|
|
7KQP
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose (P43 crystal form) | | 分子名称: | Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-11-17 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (0.88 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KR0
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, 100 K) | | 分子名称: | Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-11-18 | | 公開日 | 2020-12-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (0.77 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KR1
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, 310 K) | | 分子名称: | Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-11-18 | | 公開日 | 2020-12-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KQW
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, methylated) | | 分子名称: | Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-11-17 | | 公開日 | 2020-12-09 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (0.93 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KQO
 
 | |
5K8Z
 
 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 (pH 8.5) | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Chlorite dismutase, ... | | 著者 | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | 登録日 | 2016-05-31 | | 公開日 | 2017-06-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
7Z53
 
 | |
7QZR
 
 | |
7W6P
 
 | | Cryo-EM structure of the alpha2A adrenergic receptor GoA signaling complex bound to a G protein biased agonist | | 分子名称: | Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Xu, J, Fink, E.A, Shoichet, B.K, Du, Y. | | 登録日 | 2021-12-02 | | 公開日 | 2022-09-28 | | 最終更新日 | 2022-10-12 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | Structure-based discovery of nonopioid analgesics acting through the alpha 2A -adrenergic receptor.
Science, 377, 2022
|
|
7W7E
 
 | | Cryo-EM structure of the alpha2A adrenergic receptor GoA signaling complex bound to a biased agonist | | 分子名称: | 5-(3-bicyclo[4.2.0]octa-1,3,5-trienyl)-1,2,3,6-tetrahydropyridine, Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Xu, J, Fink, E.A, Shoichet, B.K, Du, Y. | | 登録日 | 2021-12-04 | | 公開日 | 2022-09-28 | | 最終更新日 | 2022-10-12 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure-based discovery of nonopioid analgesics acting through the alpha 2A -adrenergic receptor.
Science, 377, 2022
|
|
5MAU
 
 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 (pH 6.5) | | 分子名称: | Chlorite dismutase, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | 登録日 | 2016-11-04 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
5MFA
 
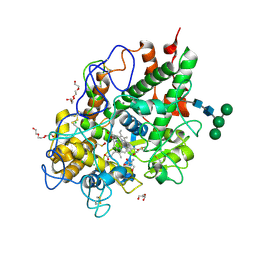 | | Crystal structure of human promyeloperoxidase (proMPO) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Grishkovskaya, I, Furtmueller, P.G, Obinger, C, Djinovic-Carugo, K. | | 登録日 | 2016-11-17 | | 公開日 | 2017-04-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structure of human promyeloperoxidase (proMPO) and the role of the propeptide in processing and maturation.
J. Biol. Chem., 292, 2017
|
|
5S4J
 
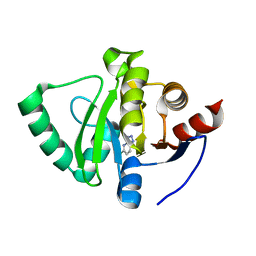 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF054 | | 分子名称: | 6-chlorotetrazolo[1,5-b]pyridazine, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.124 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S4I
 
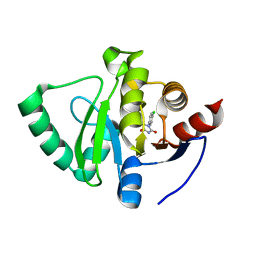 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF051 | | 分子名称: | (5S)-1-(4-chlorophenyl)-5-methylimidazolidine-2,4-dione, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.131 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S4H
 
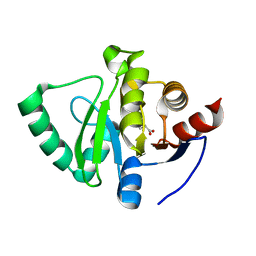 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF048 | | 分子名称: | 1-carbamoylpiperidine-4-carboxylic acid, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.175 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S4F
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF003 | | 分子名称: | 1,8-naphthyridine, Non-structural protein 3, SULFATE ION | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.131 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S4G
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF005 | | 分子名称: | Non-structural protein 3, SULFATE ION, [1,2,4]triazolo[4,3-a]pyridin-3-amine | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.172 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RSO
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000000226 | | 分子名称: | Non-structural protein 3, PARA ACETAMIDO BENZOIC ACID | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.03 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RS9
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000007636250 | | 分子名称: | 6,7-dihydro-5H-cyclopenta[d][1,2,4]triazolo[1,5-a]pyrimidin-8-amine, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RT7
 
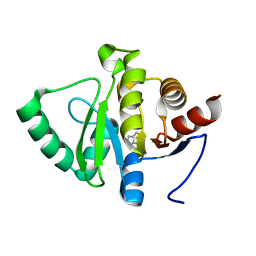 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000015442276 | | 分子名称: | 1H-PYRROLO[2,3-B]PYRIDINE, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RTO
 
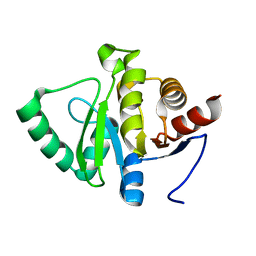 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000388302 | | 分子名称: | 4-PIPERIDINO-PIPERIDINE, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RSP
 
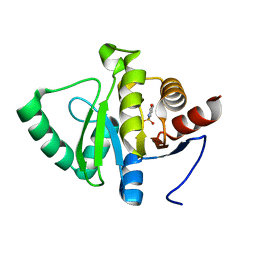 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000002560357 | | 分子名称: | 1,2-BENZISOTHIAZOL-3(2H)-ONE 1,1-DIOXIDE, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.02 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
