8I9C
 
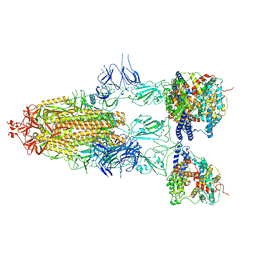 | | S-ECD (Omicron BF.7) in complex with PD of ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | 登録日 | 2023-02-06 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-08-21 | | 実験手法 | ELECTRON MICROSCOPY (3.85 Å) | | 主引用文献 | Cryo-EM structures of SARS-CoV-2 BA.2-derived subvariants spike in complex with ACE2 receptor.
Cell Discov, 9, 2023
|
|
8I9D
 
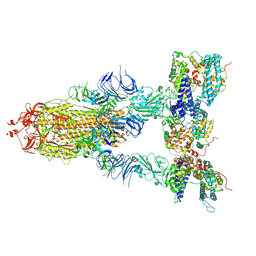 | | S-ECD (Omicron XBB.1) in complex with PD of ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | 登録日 | 2023-02-06 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-08-21 | | 実験手法 | ELECTRON MICROSCOPY (3.95 Å) | | 主引用文献 | Cryo-EM structures of SARS-CoV-2 BA.2-derived subvariants spike in complex with ACE2 receptor.
Cell Discov, 9, 2023
|
|
8I9E
 
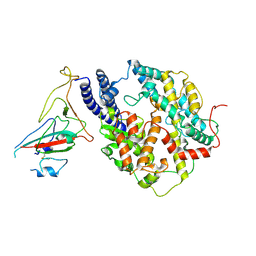 | | S-RBD(Omicron BA.3) in complex with PD of ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | 登録日 | 2023-02-06 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-08-21 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural Basis for the Enhanced Infectivity and Immune Evasion of Omicron Subvariants.
Viruses, 15, 2023
|
|
6K9C
 
 | |
8W71
 
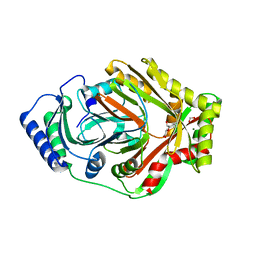 | |
7DTR
 
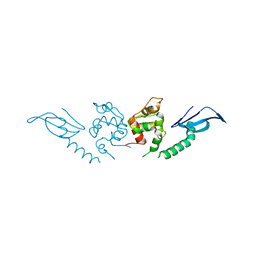 | | Structure of an AcrIF protein | | 分子名称: | AcrIF24 | | 著者 | Yue, F, Peipei, Y. | | 登録日 | 2021-01-06 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Nat Commun, 13, 2022
|
|
7Y6I
 
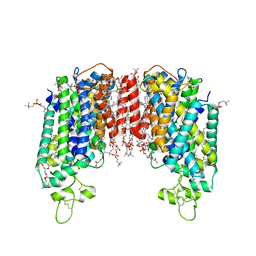 | | Cryo-EM structure of human sodium-chloride cotransporter | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Nan, J, Yang, X, Shan, Z, Yuan, Y, Zhang, Y.Q. | | 登録日 | 2022-06-20 | | 公開日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Cryo-EM structure of the human sodium-chloride cotransporter NCC.
Sci Adv, 8, 2022
|
|
7F69
 
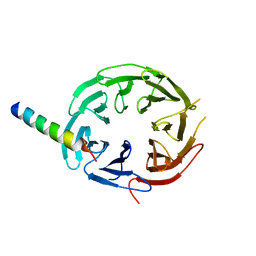 | | Crystal structure of WIPI2b in complex with ATG16L1 | | 分子名称: | Isoform 2 of Autophagy-related protein 16-1, Isoform 2 of WD repeat domain phosphoinositide-interacting protein 2 | | 著者 | Gong, X.Y, Pan, L.F. | | 登録日 | 2021-06-24 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | ATG16L1 adopts a dual-binding site mode to interact with WIPI2b in autophagy.
Sci Adv, 9, 2023
|
|
6K9E
 
 | |
8GZJ
 
 | |
8GZL
 
 | |
8GZM
 
 | |
8GZK
 
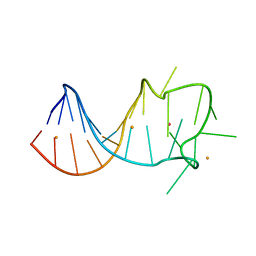 | |
8HLE
 
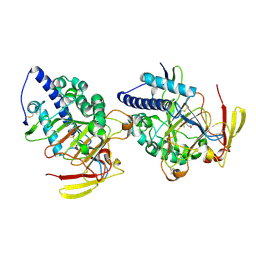 | | Structure of DddY-DMSOP complex | | 分子名称: | 3-[dimethyl(oxidanyl)-$l^{4}-sulfanyl]propanoic acid, DMSP lyase DddY, ZINC ION | | 著者 | Peng, M, Li, C.Y, Zhang, Y.Z. | | 登録日 | 2022-11-30 | | 公開日 | 2023-10-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | DMSOP-cleaving enzymes are diverse and widely distributed in marine microorganisms.
Nat Microbiol, 8, 2023
|
|
8HLF
 
 | | Crystal structure of DddK-DMSOP complex | | 分子名称: | 3-[dimethyl(oxidanyl)-$l^{4}-sulfanyl]propanoic acid, MANGANESE (II) ION, Novel protein with potential Cupin domain | | 著者 | Peng, M, Li, C.Y, Zhang, Y.Z. | | 登録日 | 2022-11-30 | | 公開日 | 2023-10-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | DMSOP-cleaving enzymes are diverse and widely distributed in marine microorganisms.
Nat Microbiol, 8, 2023
|
|
7WSG
 
 | |
7F83
 
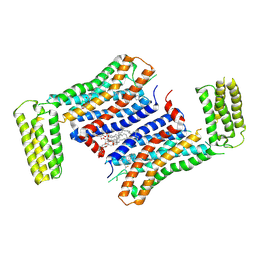 | | Crystal Structure of a receptor in Complex with inverse agonist | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(2-methylimidazo[2,1-b][1,3]thiazol-6-yl)-1-[2-[(1R)-5-(6-methylpyrimidin-4-yl)-2,3-dihydro-1H-inden-1-yl]-2,7-diazaspiro[3.5]nonan-7-yl]ethanone, Growth hormone secretagogue receptor type 1,Soluble cytochrome b562 | | 著者 | Xu, Z, Shao, Z. | | 登録日 | 2021-07-01 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | Molecular mechanism of agonism and inverse agonism in ghrelin receptor.
Nat Commun, 13, 2022
|
|
7WE6
 
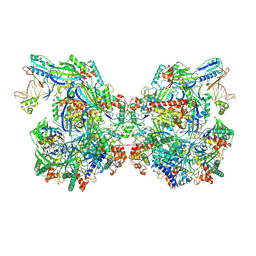 | | Structure of Csy-AcrIF24-dsDNA | | 分子名称: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | 著者 | Zhang, L, Feng, Y. | | 登録日 | 2021-12-22 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Nat Commun, 13, 2022
|
|
6JO7
 
 | | Crystal structure of mouse MXRA8 | | 分子名称: | Matrix remodeling-associated protein 8 | | 著者 | Song, H, Zhao, Z, Qi, J, Gao, F, Gao, G.F. | | 登録日 | 2019-03-20 | | 公開日 | 2019-05-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Molecular Basis of Arthritogenic Alphavirus Receptor MXRA8 Binding to Chikungunya Virus Envelope Protein.
Cell, 177, 2019
|
|
7F45
 
 | | Structure of an Anti-CRISPR protein | | 分子名称: | AcrIF5 | | 著者 | Feng, Y. | | 登録日 | 2021-06-17 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.52 Å) | | 主引用文献 | AcrIF5 specifically targets DNA-bound CRISPR-Cas surveillance complex for inhibition.
Nat.Chem.Biol., 18, 2022
|
|
7XOU
 
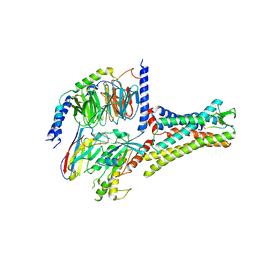 | | Structural insights into human brain gut peptide cholecystokinin receptors | | 分子名称: | CCK-8, Cholecystokinin receptor type A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Ding, Y, Zhang, H, Liao, Y, Chen, L, Ji, S. | | 登録日 | 2022-05-01 | | 公開日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural insights into human brain-gut peptide cholecystokinin receptors.
Cell Discov, 8, 2022
|
|
7XOV
 
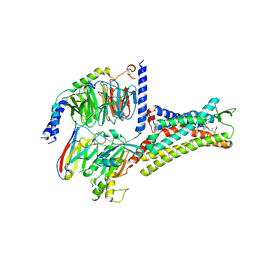 | | Structural insights into human brain gut peptide cholecystokinin receptors | | 分子名称: | 2-[2-[[4-(4-chloranyl-2,5-dimethoxy-phenyl)-5-(2-cyclohexylethyl)-1,3-thiazol-2-yl]carbamoyl]-5,7-dimethyl-indol-1-yl]ethanoic acid, Cholecystokinin receptor type A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Ding, Y, Zhang, H, Liao, Y, Chen, L, Ji, S. | | 登録日 | 2022-05-01 | | 公開日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural insights into human brain-gut peptide cholecystokinin receptors.
Cell Discov, 8, 2022
|
|
7XOW
 
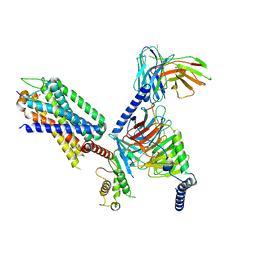 | | Structural insights into human brain gut peptide cholecystokinin receptors | | 分子名称: | Gastrin, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Ding, Y, Zhang, H, Liao, Y, Chen, L, Ji, S. | | 登録日 | 2022-05-01 | | 公開日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural insights into human brain-gut peptide cholecystokinin receptors.
Cell Discov, 8, 2022
|
|
7V8E
 
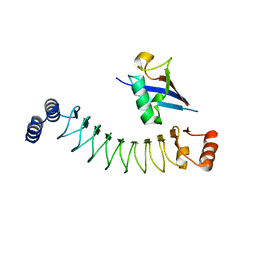 | |
7V8G
 
 | |
