7D46
 
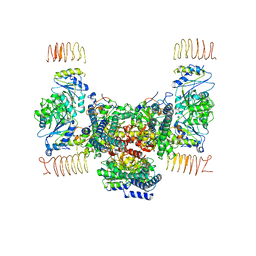 | | eIF2B apo | | 分子名称: | Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, Translation initiation factor eIF-2B subunit delta, ... | | 著者 | Kashiwagi, K, Ito, T. | | 登録日 | 2020-09-22 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | ISRIB Blunts the Integrated Stress Response by Allosterically Antagonising the Inhibitory Effect of Phosphorylated eIF2 on eIF2B.
Mol.Cell, 81, 2021
|
|
7VLK
 
 | | eIF2B-SFSV NSs C2-imposed | | 分子名称: | Non-structural protein NS-S, Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, ... | | 著者 | Kashiwagi, K, Ito, T. | | 登録日 | 2021-10-04 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.27 Å) | | 主引用文献 | eIF2B-capturing viral protein NSs suppresses the integrated stress response.
Nat Commun, 12, 2021
|
|
1WD5
 
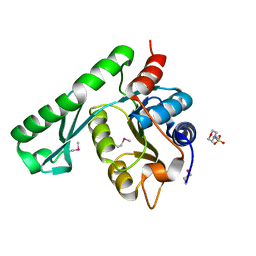 | | Crystal structure of TT1426 from Thermus thermophilus HB8 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, hypothetical protein TT1426 | | 著者 | Shibata, R, Kukimoto-Niino, M, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-05-11 | | 公開日 | 2004-11-11 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of a predicted phosphoribosyltransferase (TT1426) from Thermus thermophilus HB8 at 2.01 A resolution
Protein Sci., 14, 2005
|
|
1V4B
 
 | |
4ZLF
 
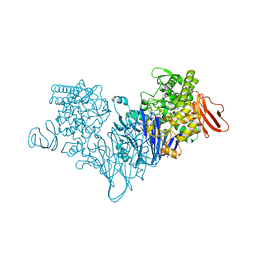 | | Cellobionic acid phosphorylase - cellobionic acid complex | | 分子名称: | 4-O-beta-D-glucopyranosyl-D-gluconic acid, CHLORIDE ION, GLYCEROL, ... | | 著者 | Nam, Y.W, Arakawa, T, Fushinobu, S. | | 登録日 | 2015-05-01 | | 公開日 | 2015-06-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
4ZLE
 
 | | Cellobionic acid phosphorylase - ligand free structure | | 分子名称: | CHLORIDE ION, GLYCEROL, Putative b-glycan phosphorylase, ... | | 著者 | Nam, Y.W, Arakawa, T, Fushinobu, S. | | 登録日 | 2015-05-01 | | 公開日 | 2015-06-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
4ZLG
 
 | | Cellobionic acid phosphorylase - gluconic acid complex | | 分子名称: | CHLORIDE ION, D-gluconic acid, D-glucono-1,5-lactone, ... | | 著者 | Nam, Y.W, Arakawa, T, Fushinobu, S. | | 登録日 | 2015-05-01 | | 公開日 | 2015-06-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
5B0S
 
 | | Beta-1,2-Mannobiose phosphorylase from Listeria innocua - beta-1,2-mannotriose complex | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Lin0857 protein, ... | | 著者 | Tsuda, T, Arakawa, T, Fushinobu, S. | | 登録日 | 2015-11-02 | | 公開日 | 2015-12-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Characterization and crystal structure determination of beta-1,2-mannobiose phosphorylase from Listeria innocua
Febs Lett., 589, 2015
|
|
4ZLI
 
 | | Cellobionic acid phosphorylase - 3-O-beta-D-glucopyranosyl-alpha-D-glucopyranuronic acid complex | | 分子名称: | CHLORIDE ION, GLYCEROL, Putative b-glycan phosphorylase, ... | | 著者 | Nam, Y.W, Arakawa, T, Fushinobu, S. | | 登録日 | 2015-05-01 | | 公開日 | 2015-06-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
7XGA
 
 | |
5B0R
 
 | | Beta-1,2-Mannobiose phosphorylase from Listeria innocua - beta-1,2-mannobiose complex | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Lin0857 protein, ... | | 著者 | Tsuda, T, Arakawa, T, Fushinobu, S. | | 登録日 | 2015-11-02 | | 公開日 | 2015-12-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Characterization and crystal structure determination of beta-1,2-mannobiose phosphorylase from Listeria innocua
Febs Lett., 589, 2015
|
|
5B0Q
 
 | | beta-1,2-Mannobiose phosphorylase from Listeria innocua - mannose complex | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lin0857 protein, SULFATE ION, ... | | 著者 | Tsuda, T, Arakawa, T, Fushinobu, S. | | 登録日 | 2015-11-02 | | 公開日 | 2015-12-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Characterization and crystal structure determination of beta-1,2-mannobiose phosphorylase from Listeria innocua
Febs Lett., 589, 2015
|
|
5B6Q
 
 | | Crystal structure of monomeric cytochrome c5 from Shewanella violacea | | 分子名称: | HEME C, IMIDAZOLE, Soluble cytochrome cA | | 著者 | Masanari, M, Fujii, S, Kawahara, K, Oki, H, Tsujino, H, Maruno, T, Kobayashi, Y, Ohkubo, T, Nishiyama, M, Harada, Y, Wakai, S, Sambongi, Y. | | 登録日 | 2016-06-01 | | 公開日 | 2016-10-19 | | 最終更新日 | 2019-10-02 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Comparative study on stabilization mechanism of monomeric cytochrome c5 from deep-sea piezophilic Shewanella violacea
Biosci.Biotechnol.Biochem., 2016
|
|
5B04
 
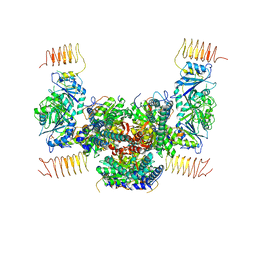 | | Crystal structure of the eukaryotic translation initiation factor 2B from Schizosaccharomyces pombe | | 分子名称: | PHOSPHATE ION, Probable translation initiation factor eIF-2B subunit beta, Probable translation initiation factor eIF-2B subunit delta, ... | | 著者 | Kashiwagi, K, Ito, T, Yokoyama, S. | | 登録日 | 2015-10-27 | | 公開日 | 2016-02-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.994 Å) | | 主引用文献 | Crystal structure of eukaryotic translation initiation factor 2B
Nature, 531, 2016
|
|
5B0P
 
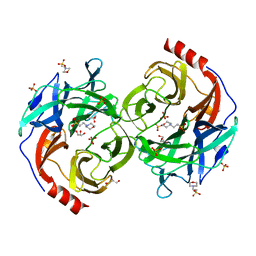 | | Beta-1,2-Mannobiose phosphorylase from Listeria innocua - glycerol complex | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Lin0857 protein, ... | | 著者 | Tsuda, T, Arakawa, T, Fushinobu, S. | | 登録日 | 2015-11-02 | | 公開日 | 2015-12-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Characterization and crystal structure determination of beta-1,2-mannobiose phosphorylase from Listeria innocua
Febs Lett., 589, 2015
|
|
1WN7
 
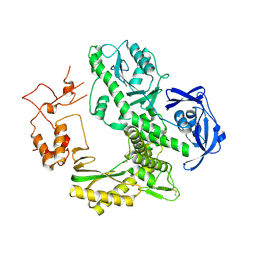 | | Crystal structure of archaeal family B DNA polymerase mutant | | 分子名称: | Family B DNA Polymerase, GLYCEROL, NICKEL (II) ION | | 著者 | Kuroita, T, Matsumura, H, Yokota, N, Hashimoto, H, Imanaka, T, Inoue, T, Kai, Y. | | 登録日 | 2004-07-28 | | 公開日 | 2005-08-02 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural Mechanism for Coordination of Proofreading and Polymerase Activities in Archaeal DNA Polymerases
J.Mol.Biol., 351, 2005
|
|
3A4T
 
 | |
1VS3
 
 | | Crystal Structure of the tRNA Pseudouridine Synthase TruA From Thermus thermophilus HB8 | | 分子名称: | tRNA pseudouridine synthase A | | 著者 | Dong, X, Bessho, Y, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-06-28 | | 公開日 | 2006-12-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal structure of tRNA pseudouridine synthase TruA from Thermus thermophilus HB8.
Rna Biol., 3, 2006
|
|
7VWS
 
 | | Carbazole Prenyl Transferase LvqB4 | | 分子名称: | 2-methyl-1-[(2R)-2-oxidanylpropyl]-9H-carbazole-3,4-dione, LvqB4, MAGNESIUM ION, ... | | 著者 | Suemune, H, Nagata, R, Kuzuyama, T, Nagano, S. | | 登録日 | 2021-11-11 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Structural Basis for the Prenylation Reaction of Carbazole-Containing Natural Products Catalyzed by Squalene Synthase-Like Enzymes.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7VWT
 
 | | Carbazole Prenyl Transferase CqsB4 | | 分子名称: | 2-methyl-1-[(2R)-2-oxidanylpropyl]-9H-carbazole-3,4-dione, CqsB4, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Suemune, H, Nagata, R, Kuzuyama, T, Nagano, S. | | 登録日 | 2021-11-11 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Structural Basis for the Prenylation Reaction of Carbazole-Containing Natural Products Catalyzed by Squalene Synthase-Like Enzymes.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7XGP
 
 | | Human renin in complex with compound3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin, UNKNOWN LIGAND | | 著者 | Kashima, A. | | 登録日 | 2022-04-05 | | 公開日 | 2022-09-07 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Discovery of SPH3127: A Novel, Highly Potent, and Orally Active Direct Renin Inhibitor.
J.Med.Chem., 65, 2022
|
|
3AJD
 
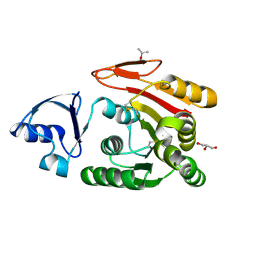 | | Crystal structure of ATRM4 | | 分子名称: | GLYCEROL, ISOPROPYL ALCOHOL, Putative methyltransferase MJ0026 | | 著者 | Hirano, M, Kuratani, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2010-06-01 | | 公開日 | 2010-06-30 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | Crystal structure of Methanocaldococcus jannaschii Trm4 complexed with sinefungin.
J.Mol.Biol., 401, 2010
|
|
3VSM
 
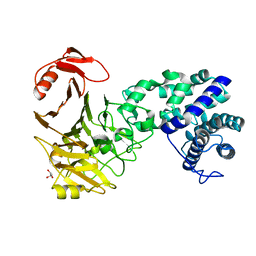 | | The crystal structure of novel chondroition lyase ODV-E66, baculovirus envelope protein | | 分子名称: | GLYCEROL, Occlusion-derived virus envelope protein E66 | | 著者 | Kawaguchi, Y, Sugiura, N, Kimata, K, Kimura, M, Kakuta, Y. | | 登録日 | 2012-04-27 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The crystal structure of novel chondroition lyase ODV-E66, baculovirus envelope protein
To be Published
|
|
3VSN
 
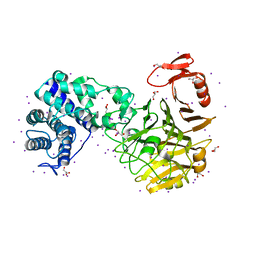 | | The crystal structure of novel chondroition lyase ODV-E66, baculovirus envelope protein | | 分子名称: | GLYCEROL, IODIDE ION, Occlusion-derived virus envelope protein E66 | | 著者 | Kawaguchi, Y, Sugiura, N, Kimata, K, Kimura, M, Kakuta, Y. | | 登録日 | 2012-04-27 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The crystal structure of novel chondroition lyase ODV-E66, baculovirus envelope protein
To be Published
|
|
3W1D
 
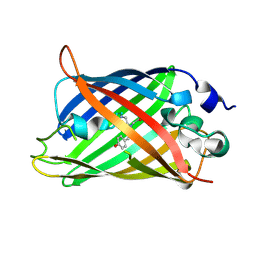 | |
