8H6D
 
 | |
8H6C
 
 | |
8H65
 
 | |
4R1S
 
 | | Crystal structure of Petunia hydrida cinnamoyl-CoA reductase | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, cinnamoyl CoA reductase | | 著者 | Noel, J.P, Louie, G.V, Bowman, M.E, Bomati, E.K. | | 登録日 | 2014-08-07 | | 公開日 | 2014-10-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural Studies of Cinnamoyl-CoA Reductase and Cinnamyl-Alcohol Dehydrogenase, Key Enzymes of Monolignol Biosynthesis.
Plant Cell, 26, 2014
|
|
8IYX
 
 | | Cryo-EM structure of the GPR34 receptor in complex with the antagonist YL-365 | | 分子名称: | 1-[4-(3-chlorophenyl)phenyl]carbonyl-4-[2-(4-phenylmethoxyphenyl)ethanoylamino]piperidine-4-carboxylic acid, Probable G-protein coupled receptor 34,YL-365 | | 著者 | Jia, G.W, Wang, X, Zhang, C.B, Dong, H.H, Su, Z.M. | | 登録日 | 2023-04-06 | | 公開日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.34 Å) | | 主引用文献 | Cryo-EM structures of human GPR34 enable the identification of selective antagonists.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4R1T
 
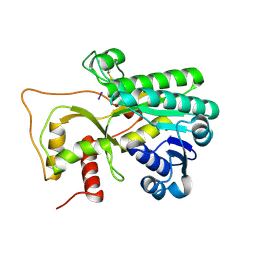 | | Crystal structure of Petunia hydrida cinnamoyl-CoA reductase | | 分子名称: | cinnamoyl CoA reductase, molecular iodine | | 著者 | Noel, J.P, Louie, G.V, Bowman, M.E, Bomati, E.K. | | 登録日 | 2014-08-07 | | 公開日 | 2014-10-01 | | 最終更新日 | 2014-11-12 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural Studies of Cinnamoyl-CoA Reductase and Cinnamyl-Alcohol Dehydrogenase, Key Enzymes of Monolignol Biosynthesis.
Plant Cell, 26, 2014
|
|
7VNU
 
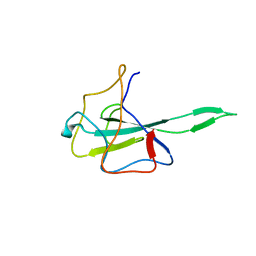 | |
5UG9
 
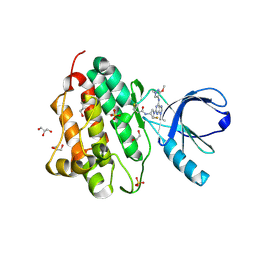 | | Crystal structure of the EGFR kinase domain (L858R, T790M, V948R) in complex with a covalent inhibitor N-[(3R,4R)-4-fluoro-1-{6-[(3-methoxy-1-methyl-1H-pyrazol-4-yl)amino]-9-(propan-2-yl)-9H-purin-2-yl}pyrrolidin-3-yl]propanamide | | 分子名称: | 1,2-ETHANEDIOL, Epidermal growth factor receptor, GLYCEROL, ... | | 著者 | Gajiwala, K.S, Ferre, R.A. | | 登録日 | 2017-01-07 | | 公開日 | 2017-03-22 | | 最終更新日 | 2018-10-10 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Discovery of N-((3R,4R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1H-pyrazol-4-yl)amino)-9-methyl-9H-purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants with Selectivity over Wild-Type EGFR.
J. Med. Chem., 60, 2017
|
|
5UGC
 
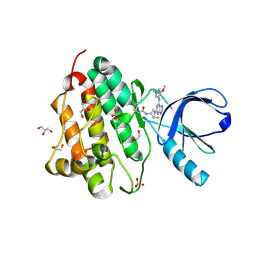 | | Crystal structure of the EGFR kinase domain (L858R, T790M, V948R) in complex with a covalent inhibitor N-[(3R,4R)-4-fluoro-1-{6-[(3-methoxy-1-methyl-1H-pyrazol-4-yl)amino]-9-methyl-9H-purin-2-yl}pyrrolidin-3-yl]propanamide | | 分子名称: | 1,2-ETHANEDIOL, Epidermal growth factor receptor, GLYCEROL, ... | | 著者 | Gajiwala, K.S, Ferre, R.A. | | 登録日 | 2017-01-08 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Discovery of N-((3R,4R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1H-pyrazol-4-yl)amino)-9-methyl-9H-purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants with Selectivity over Wild-Type EGFR.
J. Med. Chem., 60, 2017
|
|
2HAI
 
 | |
5UG8
 
 | | Crystal structure of the EGFR kinase domain (L858R, T790M, V948R) in complex with a covalent inhibitor N-[(3R,4R)-4-fluoro-1-{6-[(1-methyl-1H-pyrazol-4-yl)amino]-9-(propan-2-yl)-9H-purin-2-yl}pyrrolidin-3-yl]propanamide | | 分子名称: | Epidermal growth factor receptor, GLYCEROL, N-[(3R,4R)-4-fluoro-1-{6-[(1-methyl-1H-pyrazol-4-yl)amino]-9-(propan-2-yl)-9H-purin-2-yl}pyrrolidin-3-yl]propanamide, ... | | 著者 | Gajiwala, K.S, Ferre, R.A. | | 登録日 | 2017-01-07 | | 公開日 | 2017-03-22 | | 最終更新日 | 2017-04-26 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Discovery of N-((3R,4R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1H-pyrazol-4-yl)amino)-9-methyl-9H-purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants with Selectivity over Wild-Type EGFR.
J. Med. Chem., 60, 2017
|
|
6CAS
 
 | |
6CAR
 
 | | Serial Femtosecond X-ray Crystal Structure of 30S ribosomal subunit from Thermus thermophilus in complex with Sisomicin | | 分子名称: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, 16S Ribosomal RNA rRNA, 30S ribosomal protein S10, ... | | 著者 | DeMirci, H. | | 登録日 | 2018-01-31 | | 公開日 | 2018-07-25 | | 最終更新日 | 2018-10-24 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Aminoglycoside ribosome interactions reveal novel conformational states at ambient temperature.
Nucleic Acids Res., 46, 2018
|
|
7WZO
 
 | |
5UGA
 
 | | Crystal structure of the EGFR kinase domain (L858R, T790M, V948R) in complex with 4-(4-{[2-{[(3S)-1-acetylpyrrolidin-3-yl]amino}-9-(propan-2-yl)-9H-purin-6-yl]amino}phenyl)-1-methylpiperazin-1-ium | | 分子名称: | 4-(4-{[2-{[(3S)-1-acetylpyrrolidin-3-yl]amino}-9-(propan-2-yl)-9H-purin-6-yl]amino}phenyl)-1-methylpiperazin-1-ium, Epidermal growth factor receptor, GLYCEROL, ... | | 著者 | Gajiwala, K.S, Ferre, R.A. | | 登録日 | 2017-01-07 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Discovery of N-((3R,4R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1H-pyrazol-4-yl)amino)-9-methyl-9H-purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants with Selectivity over Wild-Type EGFR.
J. Med. Chem., 60, 2017
|
|
5UGB
 
 | |
7WM0
 
 | |
4QUK
 
 | |
4R1U
 
 | | Crystal structure of Medicago truncatula cinnamoyl-CoA reductase | | 分子名称: | ACETATE ION, Cinnamoyl CoA reductase | | 著者 | Noel, J.P, Bomati, E.K, Louie, G.V, Bowman, M.E. | | 登録日 | 2014-08-07 | | 公開日 | 2014-10-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Structural Studies of Cinnamoyl-CoA Reductase and Cinnamyl-Alcohol Dehydrogenase, Key Enzymes of Monolignol Biosynthesis.
Plant Cell, 26, 2014
|
|
7XH8
 
 | |
7WLY
 
 | | Cryo-EM structure of the Omicron S in complex with 35B5 Fab(1 down- and 2 up RBDs) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, ... | | 著者 | Wang, X, Zhu, Y. | | 登録日 | 2022-01-14 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | 35B5 antibody potently neutralizes SARS-CoV-2 Omicron by disrupting the N-glycan switch via a conserved spike epitope.
Cell Host Microbe, 30, 2022
|
|
7WLZ
 
 | |
4QTZ
 
 | |
7YKJ
 
 | | Omicron RBDs bound with P3E6 Fab (one up and one down) | | 分子名称: | P3E6 heavy chain, P3E6 light chain, Spike glycoprotein | | 著者 | Tang, B, Dang, S. | | 登録日 | 2022-07-22 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-01-18 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural insights into broadly neutralizing antibodies elicited by hybrid immunity against SARS-CoV-2.
Emerg Microbes Infect, 12, 2023
|
|
5S9O
 
 | | CRYSTAL STRUCTURE OF THE HUMAN BRD2 BD1 BROMODOMAIN IN COMPLEX WITH 9-(cyclopropylmethyl)-7-[(2R,6S)-2,6-dimethylmorpholine-4-carbonyl]-3-(3,5-dimethyl-1,2-oxazol-4-yl)-9H-carbazole-1-carboxamide | | 分子名称: | 1,2-ETHANEDIOL, 9-(cyclopropylmethyl)-7-[(2R,6S)-2,6-dimethylmorpholine-4-carbonyl]-3-(3,5-dimethyl-1,2-oxazol-4-yl)-9H-carbazole-1-carboxamide, Bromodomain-containing protein 2 | | 著者 | Sheriff, S. | | 登録日 | 2021-04-01 | | 公開日 | 2021-09-29 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Discovery and Preclinical Pharmacology of an Oral Bromodomain and Extra-Terminal (BET) Inhibitor Using Scaffold-Hopping and Structure-Guided Drug Design.
J.Med.Chem., 64, 2021
|
|
