7Y61
 
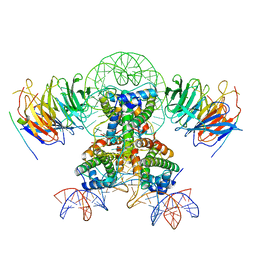 | | Cryo-EM structure of the two CAF1LCs bound right-handed Di-tetrasome | | 分子名称: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | 著者 | Liu, C.P, Yu, Z.Y, Yu, C, Xu, R.M. | | 登録日 | 2022-06-18 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | ELECTRON MICROSCOPY (5.6 Å) | | 主引用文献 | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y5V
 
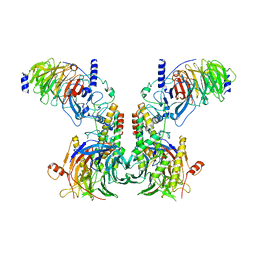 | | Cryo-EM structure of the dimeric human CAF1LC-H3-H4 complex | | 分子名称: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | 著者 | Liu, C.P, Yu, C, Yu, Z.Y, Xu, R.M. | | 登録日 | 2022-06-17 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | ELECTRON MICROSCOPY (6.1 Å) | | 主引用文献 | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y5O
 
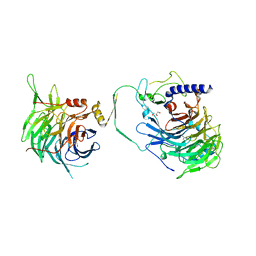 | | Crystal structure of human CAF-1 core complex in spacegroup P21 | | 分子名称: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, GLYCEROL, ... | | 著者 | Liu, C.P, Wang, M.Z, Xu, R.M. | | 登録日 | 2022-06-17 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.57 Å) | | 主引用文献 | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y5U
 
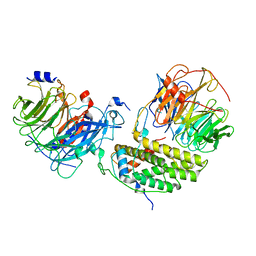 | | Cryo-EM structure of the monomeric human CAF1LC-H3-H4 complex | | 分子名称: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | 著者 | Liu, C.P, Yu, Z.Y, Yu, C, Xu, R.M. | | 登録日 | 2022-06-17 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y5W
 
 | | Cryo-EM structure of the left-handed Di-tetrasome | | 分子名称: | Histone H3.1, Histone H4, Widom 601 DNA (147-MER) | | 著者 | Liu, C.P, Yu, Z.Y, Yu, C, Xu, R.M. | | 登録日 | 2022-06-17 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y60
 
 | | Cryo-EM structure of human CAF1LC bound right-handed Di-tetrasome | | 分子名称: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | 著者 | Liu, C.P, Yu, C, Yu, Z.Y, Xu, R.M. | | 登録日 | 2022-06-18 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y5L
 
 | | Crystal structure of human CAF-1 core complex in spacegroup C2 | | 分子名称: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, GLYCEROL, ... | | 著者 | Liu, C.P, Wang, M.Z, Xu, R.M. | | 登録日 | 2022-06-17 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.42 Å) | | 主引用文献 | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7EO6
 
 | | X-ray structure analysis of xylanase | | 分子名称: | Endo-1,4-beta-xylanase, IODIDE ION | | 著者 | Wan, Q, Yi, Y, Xu, S. | | 登録日 | 2021-04-21 | | 公開日 | 2021-10-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Characterization and structural analysis of a thermophilic GH11 xylanase from compost metatranscriptome.
Appl.Microbiol.Biotechnol., 105, 2021
|
|
7EO3
 
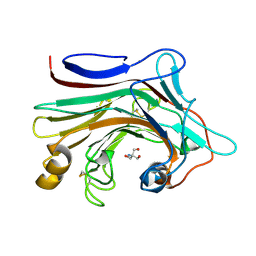 | | X-ray structure analysis of beita-1,3-glucanase | | 分子名称: | 1,3-beta-glucanase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION | | 著者 | Wan, Q, Feng, J, Xu, S. | | 登録日 | 2021-04-21 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.141 Å) | | 主引用文献 | Identification and structural analysis of a thermophilic beta-1,3-glucanase from compost
Bioresour Bioprocess, 8, 2021
|
|
7FEF
 
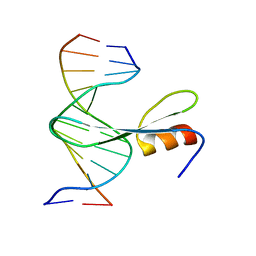 | | Crystal structure of AtMBD6 with DNA | | 分子名称: | DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain-containing protein 6 | | 著者 | Wu, Z.B, Liu, K, Min, J.R. | | 登録日 | 2021-07-19 | | 公開日 | 2021-12-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Family-wide Characterization of Methylated DNA Binding Ability of Arabidopsis MBDs.
J.Mol.Biol., 434, 2022
|
|
7FEO
 
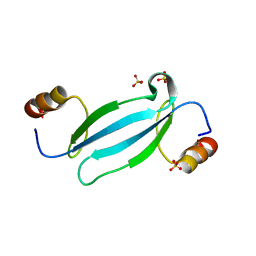 | | Crystal structure of AtMBD5 MBD domain | | 分子名称: | Methyl-CpG-binding domain-containing protein 5, SULFATE ION | | 著者 | Zhou, M.Q, Wu, Z.B, Liu, K, Min, J.R, Structural Genomics Consortium (SGC) | | 登録日 | 2021-07-21 | | 公開日 | 2021-12-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Family-wide Characterization of Methylated DNA Binding Ability of Arabidopsis MBDs.
J.Mol.Biol., 434, 2022
|
|
7F32
 
 | |
6KUU
 
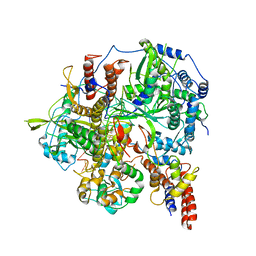 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B3) | | 分子名称: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | 著者 | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | 登録日 | 2019-09-02 | | 公開日 | 2019-12-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B3)
To Be Published
|
|
7XUZ
 
 | | Crystal structure of a HDAC4-MEF2A-DNA ternary complex | | 分子名称: | DNA (5'-D(*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*T)-3'), DNA (5'-D(P*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*A)-3'), Histone deacetylase 4, ... | | 著者 | Dai, S.Y, Guo, L, Dey, R, Guo, M, Bates, D, Cayford, J, Chen, X.J, Wei, X.D, Chen, L, Chen, Y.H. | | 登録日 | 2022-05-20 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (3.591 Å) | | 主引用文献 | Structural insights into the HDAC4-MEF2A-DNA complex and its implication in long-range transcriptional regulation.
Nucleic Acids Res., 52, 2024
|
|
7V8I
 
 | | LolCD(E171Q)E with bound AMPPNP in nanodiscs | | 分子名称: | Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolC, Lipoprotein-releasing system transmembrane protein LolE, ... | | 著者 | Bei, W.W, Luo, Q.S, Shi, H.G, Zhang, X.Z, Huang, Y.H. | | 登録日 | 2021-08-23 | | 公開日 | 2022-08-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structures of LolCDE reveal the molecular mechanism of bacterial lipoprotein sorting in Escherichia coli.
Plos Biol., 20, 2022
|
|
7V8M
 
 | | LolCDE-apo in nanodiscs | | 分子名称: | Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolC, Lipoprotein-releasing system transmembrane protein LolE | | 著者 | Luo, Q.S, Bei, W.W, Shi, H.G, Zhang, X.Z, Huang, Y.H. | | 登録日 | 2021-08-23 | | 公開日 | 2022-08-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Cryo-EM structures of LolCDE reveal the molecular mechanism of bacterial lipoprotein sorting in Escherichia coli.
Plos Biol., 20, 2022
|
|
7V8L
 
 | | LolCDE with bound RcsF in nanodiscs | | 分子名称: | (2R)-3-{[(2S)-3-HYDROXY-2-(PALMITOYLAMINO)PROPYL]THIO}PROPANE-1,2-DIYL DIHEXADECANOATE, Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolC, ... | | 著者 | Bei, W.W, Luo, Q.S, Shi, H.G, Zhang, X.Z, Huang, Y.H. | | 登録日 | 2021-08-23 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM structures of LolCDE reveal the molecular mechanism of bacterial lipoprotein sorting in Escherichia coli.
Plos Biol., 20, 2022
|
|
7YEZ
 
 | | In situ structure of polymerase complex of mammalian reovirus in the reloaded state | | 分子名称: | Lambda-2 protein, Mu-2 protein, RNA helicase, ... | | 著者 | Bao, K.Y, Zhang, X.L, Li, D.Y, Zhu, P. | | 登録日 | 2022-07-06 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | In situ structures of polymerase complex of mammalian reovirus illuminate RdRp activation and transcription regulation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YFE
 
 | | In situ structure of polymerase complex of mammalian reovirus in virion | | 分子名称: | Lambda-2 protein, Mu-2 protein, RNA (5'-R(P*AP*CP*GP*AP*UP*UP*AP*GP*C)-3'), ... | | 著者 | Bao, K.Y, Zhang, X.L, Li, D.Y, Zhu, P. | | 登録日 | 2022-07-08 | | 公開日 | 2023-03-29 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | In situ structures of polymerase complex of mammalian reovirus illuminate RdRp activation and transcription regulation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YF0
 
 | | In situ structure of polymerase complex of mammalian reovirus in the core | | 分子名称: | Lambda-2 protein, Mu-2 protein, RNA helicase, ... | | 著者 | Bao, K.Y, Zhang, X.L, Li, D.Y, Zhu, P. | | 登録日 | 2022-07-07 | | 公開日 | 2023-03-29 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | In situ structures of polymerase complex of mammalian reovirus illuminate RdRp activation and transcription regulation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YEV
 
 | | In situ structure of polymerase complex of mammalian reovirus in the pre-elongation state | | 分子名称: | Lambda-2 protein, Mu-2 protein, RNA helicase, ... | | 著者 | Bao, K.Y, Zhang, X.L, Li, D.Y, Zhu, P. | | 登録日 | 2022-07-06 | | 公開日 | 2023-03-29 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | In situ structures of polymerase complex of mammalian reovirus illuminate RdRp activation and transcription regulation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YED
 
 | | In situ structure of polymerase complex of mammalian reovirus in the elongation state | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, Lambda-2 protein, MAGNESIUM ION, ... | | 著者 | Bao, K.Y, Zhang, X.L, Li, D.Y, Zhu, P. | | 登録日 | 2022-07-05 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | In situ structures of polymerase complex of mammalian reovirus illuminate RdRp activation and transcription regulation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XM5
 
 | | Keap1 Kelch domain (residues 322-609) in complex with 6i | | 分子名称: | Kelch-like ECH-associated protein 1, N-[4-[(2-azanyl-2-oxidanylidene-ethyl)-[4-[(2-azanyl-2-oxidanylidene-ethyl)-(4-methoxyphenyl)sulfonyl-amino]naphthalen-1-yl]sulfamoyl]phenyl]-3-morpholin-4-yl-propanamide | | 著者 | Xu, K. | | 登録日 | 2022-04-24 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystallography-Guided Optimizations of the Keap1-Nrf2 Inhibitors on the Solvent Exposed Region: From Symmetric to Asymmetric Naphthalenesulfonamides.
J.Med.Chem., 65, 2022
|
|
7XM4
 
 | | Crystal structure of Keap1 Kelch domain (residues 322-609) in complex with 6e | | 分子名称: | Kelch-like ECH-associated protein 1, N-[4-[(2-azanyl-2-oxidanylidene-ethyl)-[4-[(2-azanyl-2-oxidanylidene-ethyl)-(4-methoxyphenyl)sulfonyl-amino]naphthalen-1-yl]sulfamoyl]phenyl]-2-(4-ethylpiperazin-1-yl)ethanamide | | 著者 | Xu, K. | | 登録日 | 2022-04-24 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystallography-Guided Optimizations of the Keap1-Nrf2 Inhibitors on the Solvent Exposed Region: From Symmetric to Asymmetric Naphthalenesulfonamides.
J.Med.Chem., 65, 2022
|
|
7XM3
 
 | | Crystal structure of Keap1 Kelch domain (residues 322-609) in complex with 6k | | 分子名称: | Kelch-like ECH-associated protein 1, N-[4-[(2-azanyl-2-oxidanylidene-ethyl)-[4-[(2-azanyl-2-oxidanylidene-ethyl)-(4-methoxyphenyl)sulfonyl-amino]naphthalen-1-yl]sulfamoyl]phenyl]-3-(4-ethylpiperazin-1-yl)propanamide | | 著者 | Xu, K. | | 登録日 | 2022-04-24 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystallography-Guided Optimizations of the Keap1-Nrf2 Inhibitors on the Solvent Exposed Region: From Symmetric to Asymmetric Naphthalenesulfonamides.
J.Med.Chem., 65, 2022
|
|
