8JH4
 
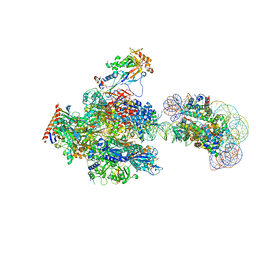 | | RNA polymerase II elongation complex containing 60 bp upstream DNA loop, stalled at SHL(-1) of the nucleosome | | 分子名称: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Akatsu, M, Fujita, R, Ogasawara, M, Ehara, H, Kujirai, T, Takizawa, Y, Sekine, S, Kurumizaka, H. | | 登録日 | 2023-05-22 | | 公開日 | 2023-11-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA.
J.Biol.Chem., 299, 2023
|
|
8JH3
 
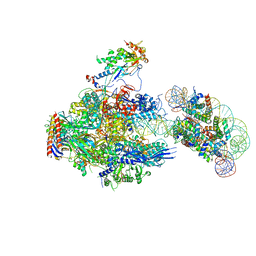 | | RNA polymerase II elongation complex containing 40 bp upstream DNA loop, stalled at SHL(-1) of the nucleosome | | 分子名称: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Akatsu, M, Fujita, R, Ogasawara, M, Ehara, H, Kujirai, T, Takizawa, Y, Sekine, S, Kurumizaka, H. | | 登録日 | 2023-05-22 | | 公開日 | 2023-11-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA.
J.Biol.Chem., 299, 2023
|
|
5CPK
 
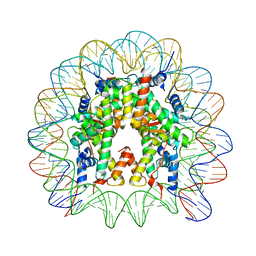 | | Nucleosome containing methylated Sat2L DNA | | 分子名称: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Osakabe, A, Arimura, Y, Adachi, F, Maehara, K, Ohkawa, Y, Kurumizaka, H. | | 登録日 | 2015-07-21 | | 公開日 | 2015-10-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.632 Å) | | 主引用文献 | Influence of DNA methylation on positioning and DNA flexibility of nucleosomes with pericentric satellite DNA.
Open Biology, 5, 2015
|
|
5CPJ
 
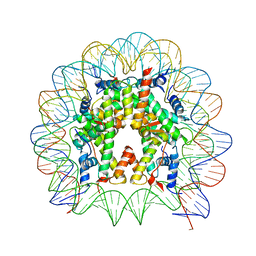 | | Nucleosome containing methylated Sat2R DNA | | 分子名称: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Osakabe, A, Arimura, Y, Adachi, F, Maehara, K, Ohkawa, Y, Kurumizaka, H. | | 登録日 | 2015-07-21 | | 公開日 | 2015-10-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Influence of DNA methylation on positioning and DNA flexibility of nucleosomes with pericentric satellite DNA.
Open Biology, 5, 2015
|
|
5CPI
 
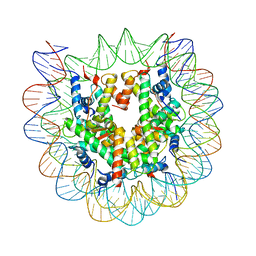 | | Nucleosome containing unmethylated Sat2R DNA | | 分子名称: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Osakabe, A, Arimura, Y, Adachi, F, Maehara, K, Ohkawa, Y, Kurumizaka, H. | | 登録日 | 2015-07-21 | | 公開日 | 2015-10-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.902 Å) | | 主引用文献 | Influence of DNA methylation on positioning and DNA flexibility of nucleosomes with pericentric satellite DNA.
Open Biology, 5, 2015
|
|
1LK5
 
 | | Structure of the D-Ribose-5-Phosphate Isomerase from Pyrococcus horikoshii | | 分子名称: | CHLORIDE ION, D-Ribose-5-Phosphate Isomerase, SODIUM ION | | 著者 | Ishikawa, K, Matsui, I, Payan, F, Cambillau, C, Ishida, H, Kawarabayasi, Y, Kikuchi, H, Roussel, A. | | 登録日 | 2002-04-24 | | 公開日 | 2002-07-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | A hyperthermostable D-ribose-5-phosphate isomerase from Pyrococcus horikoshii characterization and three-dimensional structure.
Structure, 10, 2002
|
|
1LK7
 
 | | Structure of D-Ribose-5-Phosphate Isomerase from in complex with phospho-erythronic acid | | 分子名称: | CHLORIDE ION, D-4-PHOSPHOERYTHRONIC ACID, D-Ribose-5-Phosphate Isomerase, ... | | 著者 | Ishikawa, K, Matsui, I, Payan, F, Cambillau, C, Ishida, H, Kawarabayasi, Y, Kikuchi, H, Roussel, A. | | 登録日 | 2002-04-24 | | 公開日 | 2002-07-03 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A Hyperthermostable D-Ribose-5-Phosphate Isomerase from Pyrococcus horikoshii Characterization and Three-Dimensional Structure
STRUCTURE, 10, 2002
|
|
3V10
 
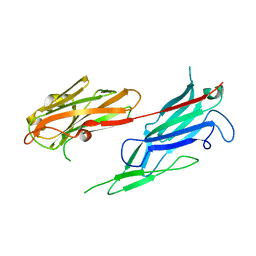 | | Crystal structure of the collagen binding domain of Erysipelothrix rhusiopathiae surface protein RspB | | 分子名称: | Rhusiopathiae surface protein B | | 著者 | Ponnuraj, K, Swarmistha devi, A, Ogawa, Y, Shimoji, Y, Subramainan, B. | | 登録日 | 2011-12-09 | | 公開日 | 2012-10-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Collagen adhesin-nanoparticle interaction impairs adhesin's ligand binding mechanism
Biochim.Biophys.Acta, 1820, 2012
|
|
3VHE
 
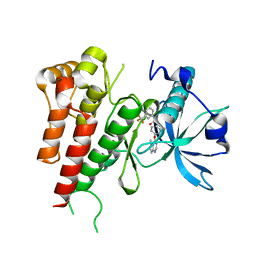 | | Crystal structure of human VEGFR2 kinase domain with a novel pyrrolopyrimidine inhibitor. | | 分子名称: | 1-{2-fluoro-4-[(5-methyl-5H-pyrrolo[3,2-d]pyrimidin-4-yl)oxy]phenyl}-3-[3-(trifluoromethyl)phenyl]urea, Vascular endothelial growth factor receptor 2 | | 著者 | Oguro, Y, Miyamoto, N, Okada, K, Takagi, T, Iwata, H, Awazu, Y, Miki, H, Hori, A, Kamiyama, K, Imanura, S. | | 登録日 | 2011-08-24 | | 公開日 | 2011-11-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Design, synthesis, and evaluation of 5-methyl-4-phenoxy-5H-pyrrolo[3,2-d]pyrimidine derivatives: novel VEGFR2 kinase inhibitors binding to inactive kinase conformation.
Bioorg.Med.Chem., 18, 2010
|
|
3W2S
 
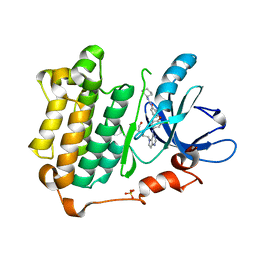 | | EGFR kinase domain with compound4 | | 分子名称: | 1-{3-[2-chloro-4-({5-[2-(2-hydroxyethoxy)ethyl]-5H-pyrrolo[3,2-d]pyrimidin-4-yl}amino)phenoxy]phenyl}-3-cyclohexylurea, Epidermal growth factor receptor, SULFATE ION | | 著者 | Sogabe, S, Kawakita, Y, Igaki, S. | | 登録日 | 2012-12-03 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-Based Approach for the Discovery of Pyrrolo[3,2-d]pyrimidine-Based EGFR T790M/L858R Mutant Inhibitors.
Acs Med.Chem.Lett., 4, 2013
|
|
3W2O
 
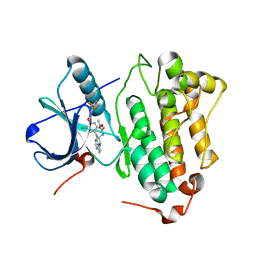 | | EGFR Kinase domain T790M/L858R Mutant with TAK-285 | | 分子名称: | Epidermal growth factor receptor, N-{2-[4-({3-chloro-4-[3-(trifluoromethyl)phenoxy]phenyl}amino)-5H-pyrrolo[3,2-d]pyrimidin-5-yl]ethyl}-3-hydroxy-3-methylbutanamide | | 著者 | Sogabe, S, Kawakita, Y, Igaki, S. | | 登録日 | 2012-12-03 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structure-Based Approach for the Discovery of Pyrrolo[3,2-d]pyrimidine-Based EGFR T790M/L858R Mutant Inhibitors.
Acs Med.Chem.Lett., 4, 2013
|
|
3W2R
 
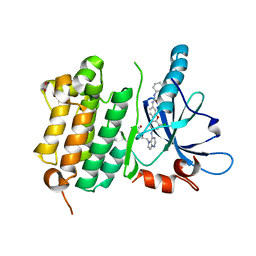 | | EGFR Kinase domain T790M/L858R mutant with compound 4 | | 分子名称: | 1,2-ETHANEDIOL, 1-{3-[2-chloro-4-({5-[2-(2-hydroxyethoxy)ethyl]-5H-pyrrolo[3,2-d]pyrimidin-4-yl}amino)phenoxy]phenyl}-3-cyclohexylurea, Epidermal growth factor receptor | | 著者 | Sogabe, S, Kawakita, Y, Igaki, S. | | 登録日 | 2012-12-03 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure-Based Approach for the Discovery of Pyrrolo[3,2-d]pyrimidine-Based EGFR T790M/L858R Mutant Inhibitors.
Acs Med.Chem.Lett., 4, 2013
|
|
3W2P
 
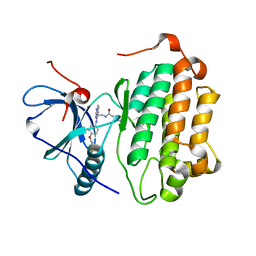 | | EGFR Kinase domain T790M/L858R mutant with compound 2 | | 分子名称: | Epidermal growth factor receptor, N-{2-[4-({3-chloro-4-[3-(trifluoromethyl)phenoxy]phenyl}amino)-5H-pyrrolo[3,2-d]pyrimidin-5-yl]ethyl}-4-(dimethylamino)butanamide | | 著者 | Sogabe, S, Kawakita, Y, Igaki, S. | | 登録日 | 2012-12-03 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure-Based Approach for the Discovery of Pyrrolo[3,2-d]pyrimidine-Based EGFR T790M/L858R Mutant Inhibitors.
Acs Med.Chem.Lett., 4, 2013
|
|
3W2Q
 
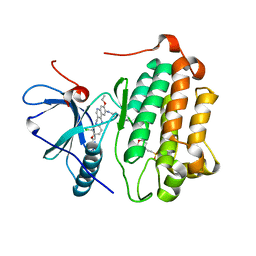 | | EGFR kinase domain T790M/L858R mutant with HKI-272 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Epidermal growth factor receptor, N-(4-{[3-chloro-4-(pyridin-2-ylmethoxy)phenyl]amino}-3-cyano-7-ethoxyquinolin-6-yl)-4-(dimethylamino)butanamide | | 著者 | Sogabe, S, Kawakita, Y, Igaki, S. | | 登録日 | 2012-12-03 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure-Based Approach for the Discovery of Pyrrolo[3,2-d]pyrimidine-Based EGFR T790M/L858R Mutant Inhibitors.
Acs Med.Chem.Lett., 4, 2013
|
|
5B0H
 
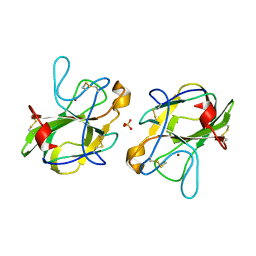 | | CRYSTAL STRUCTURE OF HUMAN LEUKOCYTE CELL-DERIVED CHEMOTAXIN 2 | | 分子名称: | Leukocyte cell-derived chemotaxin-2, SULFATE ION, ZINC ION | | 著者 | Zheng, H, Miyakawa, T, Sawano, Y, Tanokura, M. | | 登録日 | 2015-10-29 | | 公開日 | 2016-07-06 | | 最終更新日 | 2020-02-26 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Crystal Structure of Human Leukocyte Cell-derived Chemotaxin 2 (LECT2) Reveals a Mechanistic Basis of Functional Evolution in a Mammalian Protein with an M23 Metalloendopeptidase Fold
J.Biol.Chem., 291, 2016
|
|
4FRU
 
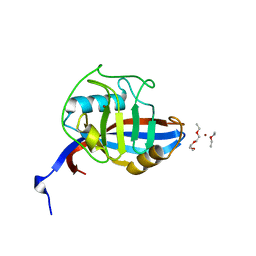 | | Crystal structure of horse wild-type cyclophilin B | | 分子名称: | 1-ETHOXY-2-(2-METHOXYETHOXY)ETHANE, DI(HYDROXYETHYL)ETHER, Peptidyl-prolyl cis-trans isomerase, ... | | 著者 | Boudko, S.P, Ishikawa, Y, Bachinger, H.P. | | 登録日 | 2012-06-26 | | 公開日 | 2012-11-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Crystal structures of wild-type and mutated cyclophilin B that causes hyperelastosis cutis in the American quarter horse.
BMC Res Notes, 5, 2012
|
|
4FRV
 
 | | Crystal structure of mutated cyclophilin B that causes hyperelastosis cutis in the American Quarter Horse | | 分子名称: | 1-ETHOXY-2-(2-METHOXYETHOXY)ETHANE, DI(HYDROXYETHYL)ETHER, Peptidyl-prolyl cis-trans isomerase, ... | | 著者 | Boudko, S.P, Ishikawa, Y, Bachinger, H.P. | | 登録日 | 2012-06-26 | | 公開日 | 2012-11-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Crystal structures of wild-type and mutated cyclophilin B that causes hyperelastosis cutis in the American quarter horse.
BMC Res Notes, 5, 2012
|
|
3X3Y
 
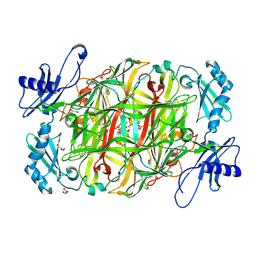 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by histamine | | 分子名称: | COPPER (II) ION, GLYCEROL, POTASSIUM ION, ... | | 著者 | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | 登録日 | 2015-03-10 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.499 Å) | | 主引用文献 | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3X42
 
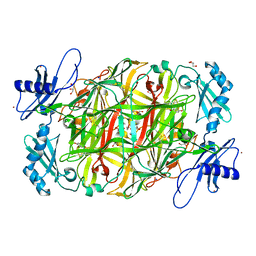 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis in the presence of sodium bromide | | 分子名称: | BROMIDE ION, COPPER (II) ION, GLYCEROL, ... | | 著者 | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | 登録日 | 2015-03-10 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.875 Å) | | 主引用文献 | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3X41
 
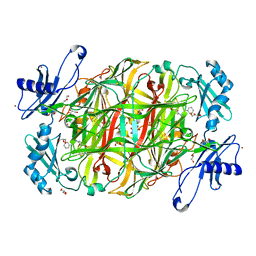 | | Copper amine oxidase from Arthrobacter globiformis: Product Schiff-base form produced by anaerobic reduction in the presence of sodium bromide | | 分子名称: | BROMIDE ION, COPPER (II) ION, GLYCEROL, ... | | 著者 | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | 登録日 | 2015-03-10 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3X3X
 
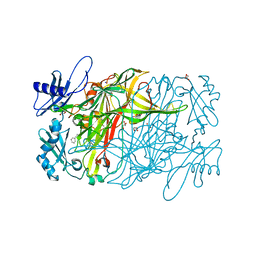 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by phenylethylamine | | 分子名称: | 2-PHENYL-ETHANOL, COPPER (II) ION, GLYCEROL, ... | | 著者 | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | 登録日 | 2015-03-10 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3X3Z
 
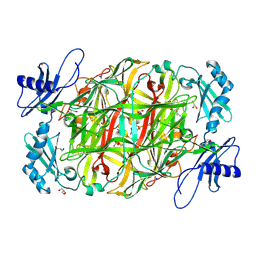 | | Copper amine oxidase from Arthrobacter globiformis: Aminoresorcinol form produced by anaerobic reduction with ethylamine hydrochloride | | 分子名称: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | 著者 | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | 登録日 | 2015-03-10 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3X40
 
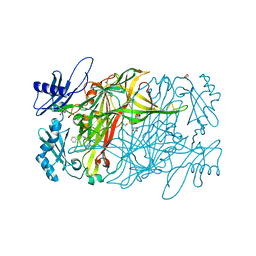 | | Copper amine oxidase from Arthrobacter globiformis: Product Schiff-base form produced by anaerobic reduction in the presence of sodium chloride | | 分子名称: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | 著者 | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | 登録日 | 2015-03-10 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
7CCO
 
 | | The binding structure of a lanthanide binding tag (LBT3) with lanthanum ion (La3+) | | 分子名称: | LANTHANUM (III) ION, LBT3 | | 著者 | Hatanaka, T, Kikkawa, N, Matsugami, A, Hosokawa, Y, Hayashi, F, Ishida, N. | | 登録日 | 2020-06-17 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The origins of binding specificity of a lanthanide ion binding peptide.
Sci Rep, 10, 2020
|
|
7CCN
 
 | | The binding structure of a lanthanide binding tag (LBT3) with lutetium ion (Lu3+) | | 分子名称: | LBT3, LUTETIUM (III) ION | | 著者 | Hatanaka, T, Kikkawa, N, Matsugami, A, Hosokawa, Y, Hayashi, F, Ishida, N. | | 登録日 | 2020-06-17 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The origins of binding specificity of a lanthanide ion binding peptide.
Sci Rep, 10, 2020
|
|
