7FDV
 
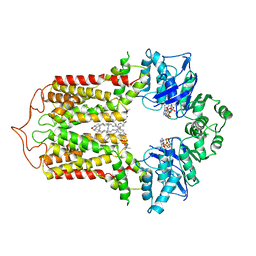 | | Cryo-EM structure of the human cholesterol transporter ABCG1 in complex with cholesterol | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family G member 1, CHOLESTEROL, ... | | Authors: | Xu, D, Li, Y.Y, Yang, F.R, Sun, C.R, Pan, J.H, Wang, L, Chen, Z.P, Fang, S.C, Yao, X.B, Hou, W.T, Zhou, C.Z, Chen, Y. | | Deposit date: | 2021-07-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure and transport mechanism of the human cholesterol transporter ABCG1.
Cell Rep, 38, 2022
|
|
3BD9
 
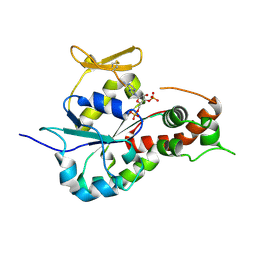 | | human 3-O-sulfotransferase isoform 5 with bound PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Heparan sulfate glucosamine 3-O-sulfotransferase 5 | | Authors: | Xu, D, Moon, A.F, Song, D, Liu, J, Pedersen, L.C. | | Deposit date: | 2007-11-14 | | Release date: | 2008-01-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Engineering sulfotransferases to modify heparan sulfate.
Nat.Chem.Biol., 4, 2008
|
|
1NWB
 
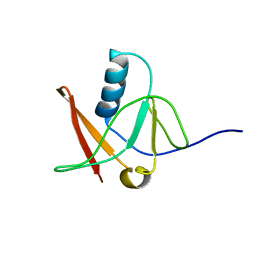 | | Solution NMR Structure of Protein AQ_1857 from Aquifex aeolicus: Northeast Structural Genomics Consortium Target QR6. | | Descriptor: | Hypothetical protein AQ_1857 | | Authors: | Xu, D, Liu, G, Xiao, R, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-05 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the hypothetical protein AQ-1857 encoded by the Y157 gene from Aquifex aeolicus reveals a novel protein fold.
Proteins, 54, 2004
|
|
9BX2
 
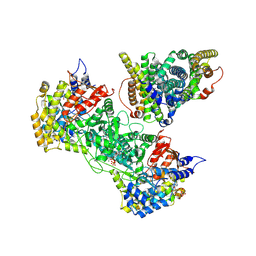 | | Class 2 model for preturnover condition of Bacillus subtilis ribonucleotide reductase complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Xu, D, Thomas, W.C, Burnim, A.A, Ando, N. | | Deposit date: | 2024-05-22 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Conformational landscapes of a class I ribonucleotide reductase complex during turnover reveal intrinsic dynamics and asymmetry.
Nat Commun, 16, 2025
|
|
9BX3
 
 | | Class 5 model for preturnover condition of Bacillus subtilis ribonucleotide reductase complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Xu, D, Thomas, W.C, Burnim, A.A, Ando, N. | | Deposit date: | 2024-05-22 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Conformational landscapes of a class I ribonucleotide reductase complex during turnover reveal intrinsic dynamics and asymmetry.
Nat Commun, 16, 2025
|
|
5GWD
 
 | | Structure of Myroilysin | | Descriptor: | Myroilysin, ZINC ION | | Authors: | Xu, D, Ran, T, Wang, W. | | Deposit date: | 2016-09-10 | | Release date: | 2017-02-15 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Myroilysin Is a New Bacterial Member of the M12A Family of Metzincin Metallopeptidases and Is Activated by a Cysteine Switch Mechanism.
J. Biol. Chem., 292, 2017
|
|
4IM8
 
 | | low resolution crystal structure of mouse RAGE | | Descriptor: | Advanced glycation end-products receptor | | Authors: | Xu, D, Young, J.H, Krahn, J.M, Song, D, Corbett, K.D, Chazin, W.J, Pedersen, L.C, Esko, J.D. | | Deposit date: | 2013-01-02 | | Release date: | 2013-08-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Stable RAGE-Heparan Sulfate Complexes Are Essential for Signal Transduction.
Acs Chem.Biol., 8, 2013
|
|
9BY9
 
 | | Class 10 model for product condition of Bacillus subtilis ribonucleotide reductase complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xu, D, Thomas, W.C, Burnim, A.A, Ando, N. | | Deposit date: | 2024-05-23 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Conformational landscapes of a class I ribonucleotide reductase complex during turnover reveal intrinsic dynamics and asymmetry.
Nat Commun, 16, 2025
|
|
9BZ5
 
 | | Class 6 model for combined refinement of Bacillus subtilis ribonucleotide reductase complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xu, D, Thomas, W.C, Burnim, A.A, Ando, N. | | Deposit date: | 2024-05-24 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Conformational landscapes of a class I ribonucleotide reductase complex during turnover reveal intrinsic dynamics and asymmetry.
Nat Commun, 16, 2025
|
|
9BXS
 
 | | Consensus full-complex model for pre-reduction condition of Bacillus subtilis ribonucleotide reductase complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xu, D, Thomas, W.C, Burnim, A.A, Ando, N. | | Deposit date: | 2024-05-22 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Conformational landscapes of a class I ribonucleotide reductase complex during turnover reveal intrinsic dynamics and asymmetry.
Nat Commun, 16, 2025
|
|
9BYX
 
 | | Class 8 model for turnover condition of Bacillus subtilis ribonucleotide reductase complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xu, D, Thomas, W.C, Burnim, A.A, Ando, N. | | Deposit date: | 2024-05-24 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Conformational landscapes of a class I ribonucleotide reductase complex during turnover reveal intrinsic dynamics and asymmetry.
Nat Commun, 16, 2025
|
|
9BZ6
 
 | | Class 7 model for combined refinement of Bacillus subtilis ribonucleotide reductase complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xu, D, Thomas, W.C, Burnim, A.A, Ando, N. | | Deposit date: | 2024-05-24 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Conformational landscapes of a class I ribonucleotide reductase complex during turnover reveal intrinsic dynamics and asymmetry.
Nat Commun, 16, 2025
|
|
9BXX
 
 | | Class 11 model for pre-reduction condition of Bacillus subtilis ribonucleotide reductase complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xu, D, Thomas, W.C, Burnim, A.A, Ando, N. | | Deposit date: | 2024-05-23 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | Conformational landscapes of a class I ribonucleotide reductase complex during turnover reveal intrinsic dynamics and asymmetry.
Nat Commun, 16, 2025
|
|
9BZK
 
 | | Class 43 model for combined refinement of Bacillus subtilis ribonucleotide reductase complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xu, D, Thomas, W.C, Burnim, A.A, Ando, N. | | Deposit date: | 2024-05-24 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Conformational landscapes of a class I ribonucleotide reductase complex during turnover reveal intrinsic dynamics and asymmetry.
Nat Commun, 16, 2025
|
|
9BY8
 
 | | Class 9 model for product condition of Bacillus subtilis ribonucleotide reductase complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xu, D, Thomas, W.C, Burnim, A.A, Ando, N. | | Deposit date: | 2024-05-23 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Conformational landscapes of a class I ribonucleotide reductase complex during turnover reveal intrinsic dynamics and asymmetry.
Nat Commun, 16, 2025
|
|
9BYZ
 
 | | Class 12 model for turnover condition of Bacillus subtilis ribonucleotide reductase complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xu, D, Thomas, W.C, Burnim, A.A, Ando, N. | | Deposit date: | 2024-05-24 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Conformational landscapes of a class I ribonucleotide reductase complex during turnover reveal intrinsic dynamics and asymmetry.
Nat Commun, 16, 2025
|
|
9BYH
 
 | | Consensus model for turnover condition of Bacillus subtilis ribonucleotide reductase complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xu, D, Thomas, W.C, Burnim, A.A, Ando, N. | | Deposit date: | 2024-05-23 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Conformational landscapes of a class I ribonucleotide reductase complex during turnover reveal intrinsic dynamics and asymmetry.
Nat Commun, 16, 2025
|
|
9BZA
 
 | | Class 18 model for combined refinement of Bacillus subtilis ribonucleotide reductase complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xu, D, Thomas, W.C, Burnim, A.A, Ando, N. | | Deposit date: | 2024-05-24 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Conformational landscapes of a class I ribonucleotide reductase complex during turnover reveal intrinsic dynamics and asymmetry.
Nat Commun, 16, 2025
|
|
9BY2
 
 | | Consensus full-complex model for product condition of Bacillus subtilis ribonucleotide reductase complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xu, D, Thomas, W.C, Burnim, A.A, Ando, N. | | Deposit date: | 2024-05-23 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Conformational landscapes of a class I ribonucleotide reductase complex during turnover reveal intrinsic dynamics and asymmetry.
Nat Commun, 16, 2025
|
|
9BYA
 
 | | Class 11 model for product condition of Bacillus subtilis ribonucleotide reductase complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xu, D, Thomas, W.C, Burnim, A.A, Ando, N. | | Deposit date: | 2024-05-23 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Conformational landscapes of a class I ribonucleotide reductase complex during turnover reveal intrinsic dynamics and asymmetry.
Nat Commun, 16, 2025
|
|
9BZ9
 
 | | Class 15 model for combined refinement of Bacillus subtilis ribonucleotide reductase complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xu, D, Thomas, W.C, Burnim, A.A, Ando, N. | | Deposit date: | 2024-05-24 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (4.64 Å) | | Cite: | Conformational landscapes of a class I ribonucleotide reductase complex during turnover reveal intrinsic dynamics and asymmetry.
Nat Commun, 16, 2025
|
|
9BYL
 
 | | Consensus full-complex model for turnover condition of Bacillus subtilis ribonucleotide reductase complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xu, D, Thomas, W.C, Burnim, A.A, Ando, N. | | Deposit date: | 2024-05-23 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Conformational landscapes of a class I ribonucleotide reductase complex during turnover reveal intrinsic dynamics and asymmetry.
Nat Commun, 16, 2025
|
|
9BZF
 
 | | Class 28 model for combined refinement of Bacillus subtilis ribonucleotide reductase complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xu, D, Thomas, W.C, Burnim, A.A, Ando, N. | | Deposit date: | 2024-05-24 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Conformational landscapes of a class I ribonucleotide reductase complex during turnover reveal intrinsic dynamics and asymmetry.
Nat Commun, 16, 2025
|
|
9BXZ
 
 | | Class 15 model for pre-reduction condition of Bacillus subtilis ribonucleotide reductase complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xu, D, Thomas, W.C, Burnim, A.A, Ando, N. | | Deposit date: | 2024-05-23 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (8.11 Å) | | Cite: | Conformational landscapes of a class I ribonucleotide reductase complex during turnover reveal intrinsic dynamics and asymmetry.
Nat Commun, 16, 2025
|
|
9BYV
 
 | | Class 4 model for turnover condition of Bacillus subtilis ribonucleotide reductase complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xu, D, Thomas, W.C, Burnim, A.A, Ando, N. | | Deposit date: | 2024-05-24 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Conformational landscapes of a class I ribonucleotide reductase complex during turnover reveal intrinsic dynamics and asymmetry.
Nat Commun, 16, 2025
|
|
