1Z7Z
 
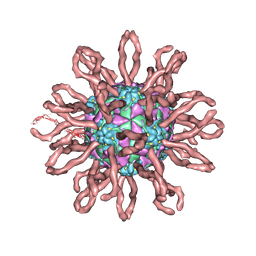 | | Cryo-em structure of human coxsackievirus A21 complexed with five domain icam-1kilifi | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Intercellular adhesion molecule-1, human coxsackievirus A21 | | Authors: | Xiao, C, Bator-Kelly, C.M, Rieder, E, Chipman, P.R, Craig, A, Kuhn, R.J, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2005-03-28 | | Release date: | 2005-08-02 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | The crystal structure of coxsackievirus a21 and its interaction with icam-1.
Structure, 13, 2005
|
|
1Z7S
 
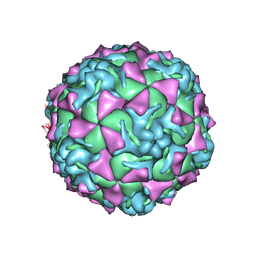 | | The crystal structure of coxsackievirus A21 | | Descriptor: | CALCIUM ION, GUANOSINE-5'-MONOPHOSPHATE, Human COXSACKIEVIRUS A21, ... | | Authors: | Xiao, C, Bator-Kelly, C.M, Rieder, E, Chipman, P.R, Craig, A, Kuhn, R.J, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2005-03-28 | | Release date: | 2005-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of coxsackievirus a21 and its interaction with icam-1.
Structure, 13, 2005
|
|
7DTL
 
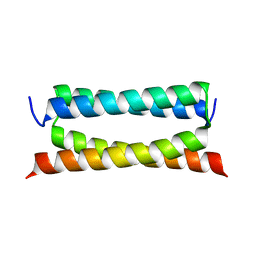 | | Crystal structure of PSK, an antimicrobial peptide from Chrysomya megacephala | | Descriptor: | PSK | | Authors: | Xiao, C, Xiao, Z, Wang, S, Liu, W. | | Deposit date: | 2021-01-05 | | Release date: | 2021-07-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal and solution structures of a novel antimicrobial peptide from Chrysomya megacephala.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5BNO
 
 | | Crystal structure of human enterovirus D68 in complex with 6'SLN | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Liu, Y, Sheng, J, Meng, G, Xiao, C, Rossmann, M.G. | | Deposit date: | 2015-05-26 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Sialic acid-dependent cell entry of human enterovirus D68.
Nat Commun, 6, 2015
|
|
5BNP
 
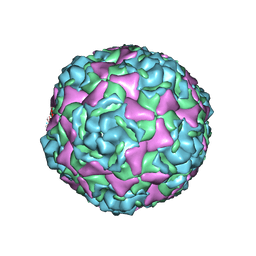 | | Crystal structure of human enterovirus D68 in complex with 3'SLN | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Liu, Y, Sheng, J, Meng, G, Xiao, C, Rossmann, M.G. | | Deposit date: | 2015-05-26 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Sialic acid-dependent cell entry of human enterovirus D68.
Nat Commun, 6, 2015
|
|
5BNN
 
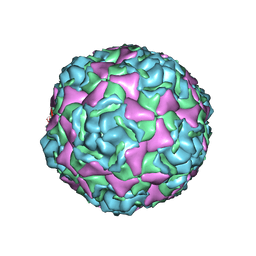 | | Crystal structure of human enterovirus D68 in complex with 6'SL | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Liu, Y, Sheng, J, Meng, G, Xiao, C, Rossmann, M.G. | | Deposit date: | 2015-05-26 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Sialic acid-dependent cell entry of human enterovirus D68.
Nat Commun, 6, 2015
|
|
7X8M
 
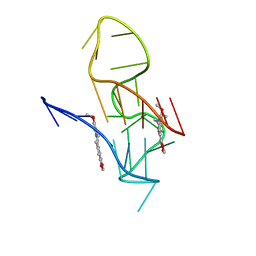 | | NMR Solution Structure of the 2:1 Berberine-KRAS-G4 Complex | | Descriptor: | BERBERINE, DNA (24-MER) | | Authors: | Wang, K.B, Liu, Y, Li, J, Xiao, C, Gu, W, Li, Y, Xia, Y.Z, Yan, T, Yang, M.H, Kong, L.Y. | | Deposit date: | 2022-03-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the bulge-containing KRAS oncogene promoter G-quadruplex bound to berberine and coptisine.
Nat Commun, 13, 2022
|
|
7X8O
 
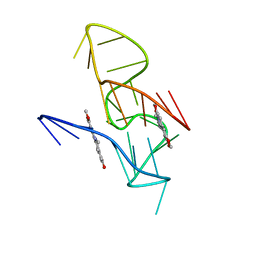 | | NMR Solution Structure of the 2:1 Coptisine-KRAS-G4 Complex | | Descriptor: | 6,7-dihydro[1,3]dioxolo[4,5-g][1,3]dioxolo[7,8]isoquino[3,2-a]isoquinolin-5-ium, DNA (24-MER) | | Authors: | Wang, K.B, Liu, Y, Li, J, Xiao, C, Gu, W, Li, Y, Xia, Y.Z, Yan, T, Yang, M.H, Kong, L.Y. | | Deposit date: | 2022-03-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the bulge-containing KRAS oncogene promoter G-quadruplex bound to berberine and coptisine.
Nat Commun, 13, 2022
|
|
7X8N
 
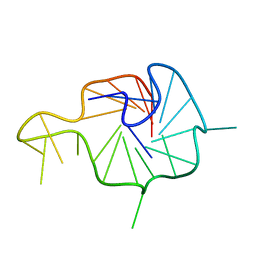 | | NMR Solution Structure of the Wild-type Bulge-containing KRAS-G4 | | Descriptor: | DNA (24-mer) | | Authors: | Wang, K.B, Liu, Y, Li, J, Xiao, C, Gu, W, Li, Y, Xia, Y.Z, Yan, T, Yang, M.H, Kong, L.Y. | | Deposit date: | 2022-03-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the bulge-containing KRAS oncogene promoter G-quadruplex bound to berberine and coptisine.
Nat Commun, 13, 2022
|
|
7MF1
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody 47D1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 47D1 Fab heavy chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2021-04-08 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Diverse immunoglobulin gene usage and convergent epitope targeting in neutralizing antibody responses to SARS-CoV-2.
Cell Rep, 35, 2021
|
|
5DH3
 
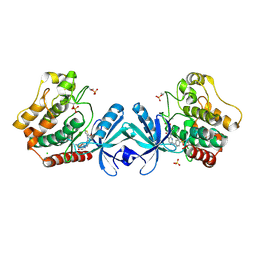 | | Crystal structure of MST2 in complex with XMU-MP-1 | | Descriptor: | 4-[(5,10-dimethyl-6-oxo-6,10-dihydro-5H-pyrimido[5,4-b]thieno[3,2-e][1,4]diazepin-2-yl)amino]benzenesulfonamide, CHLORIDE ION, SULFATE ION, ... | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2015-08-29 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.468 Å) | | Cite: | Pharmacological targeting of kinases MST1 and MST2 augments tissue repair and regeneration
Sci Transl Med, 8, 2016
|
|
2B6B
 
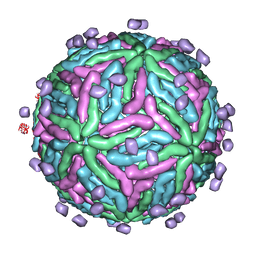 | | Cryo EM structure of Dengue complexed with CRD of DC-SIGN | | Descriptor: | CD209 antigen, envelope glycoprotein | | Authors: | Pokidysheva, E, Zhang, Y, Battisti, A.J, Bator-Kelly, C.M, Chipman, P.R, Gregorio, G, Hendrickson, W.A, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2005-09-30 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Cryo-EM reconstruction of dengue virus in complex with the carbohydrate recognition domain of DC-SIGN
Cell(Cambridge,Mass.), 124, 2006
|
|
8GPD
 
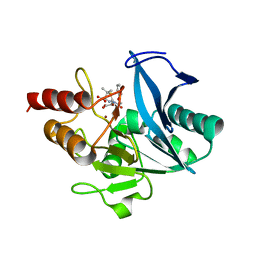 | | Crystal structure of NDM-1 at pH5.5 (Succinate) in complex with hydrolyzed penicillin V | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-2-oxidanyl-2-oxidanylidene-1-(2-phenoxyethanoylamino)ethyl]-1,3-thiazolidine-4-carboxylic acid, Metallo beta lactamase NDM-1, POTASSIUM ION, ... | | Authors: | Shi, X, Dai, Y, Zhang, Q, Liu, W. | | Deposit date: | 2022-08-26 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interplay between the beta-lactam side chain and an active-site mobile loop of NDM-1 in penicillin hydrolysis as a potential target for mechanism-based inhibitor design.
Int.J.Biol.Macromol., 262, 2024
|
|
8GPC
 
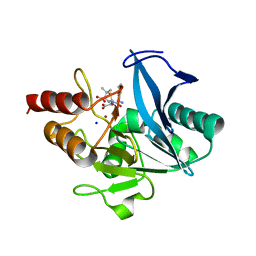 | | Crystal structure of NDM-1 at pH5.5 (Succinate) in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Metallo beta lactamase NDM-1, SODIUM ION, ... | | Authors: | Shi, X, Dai, Y, Zhang, Q, Liu, W. | | Deposit date: | 2022-08-26 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interplay between the beta-lactam side chain and an active-site mobile loop of NDM-1 in penicillin hydrolysis as a potential target for mechanism-based inhibitor design.
Int.J.Biol.Macromol., 262, 2024
|
|
8GPE
 
 | | Crystal structure of NDM-1 at pH5.5 (Succinate) in complex with hydrolyzed penicillin G | | Descriptor: | (2R,4S)-2-{(R)-carboxy[(phenylacetyl)amino]methyl}-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Metallo beta lactamase NDM-1, POTASSIUM ION, ... | | Authors: | Shi, X, Dai, Y, Zhang, Q, Liu, W. | | Deposit date: | 2022-08-26 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interplay between the beta-lactam side chain and an active-site mobile loop of NDM-1 in penicillin hydrolysis as a potential target for mechanism-based inhibitor design.
Int.J.Biol.Macromol., 262, 2024
|
|
8WA8
 
 | | Human transketolase in complex with phosphite | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PHOSPHITE ION, ... | | Authors: | Liu, Z, Tittmann, K, Dai, S. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Multifaceted Role of the Substrate Phosphate Group in Transketolase Catalysis
Acs Catalysis, 14, 2024
|
|
8WAA
 
 | | Human transketolase soaked with donor ketose D-xylulose | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-[(4-AMINO-2-METHYL-5-PYRIMIDINYL)METHYL]-2-(1,2-DIHYDROXYETHYL)-4-METHYL-1,3-THIAZOL-3-IUM-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Liu, Z, Dai, S, Tittmann, K. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Multifaceted Role of the Substrate Phosphate Group in Transketolase Catalysis
Acs Catalysis, 14, 2024
|
|
8HVS
 
 | |
8I8F
 
 | | Crystal structure of NDM-1 at pH5.5 (Succinate) in complex with hydrolyzed compound 1 | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-(2-naphthalen-1-yloxyethanoylamino)-2-oxidanyl-2-oxidanylidene-ethyl]-1,3-thiazolidine-4-carboxylic acid, Metallo beta lactamase NDM-1, ZINC ION | | Authors: | Shi, X, Liu, W. | | Deposit date: | 2023-02-04 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Interplay between the beta-lactam side chain and an active-site mobile loop of NDM-1 in penicillin hydrolysis as a potential target for mechanism-based inhibitor design.
Int.J.Biol.Macromol., 262, 2024
|
|
8WA9
 
 | | Human transketolase soaked with donor ketose D-fructose | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-[(4-AMINO-2-METHYL-5-PYRIMIDINYL)METHYL]-2-(1,2-DIHYDROXYETHYL)-4-METHYL-1,3-THIAZOL-3-IUM-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Liu, Z, Tittmann, K, Dai, S. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Multifaceted Role of the Substrate Phosphate Group in Transketolase Catalysis
Acs Catalysis, 14, 2024
|
|
8WA7
 
 | | E.coli transketolase soaked with donor ketose D-fructose | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-[(4-AMINO-2-METHYL-5-PYRIMIDINYL)METHYL]-2-(1,2-DIHYDROXYETHYL)-4-METHYL-1,3-THIAZOL-3-IUM-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, GLYCEROL, ... | | Authors: | Liu, Z, Dai, S, Tittmann, K. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Multifaceted Role of the Substrate Phosphate Group in Transketolase Catalysis
Acs Catalysis, 14, 2024
|
|
7V21
 
 | |
7V1Y
 
 | |
7V1Z
 
 | | human Serine beta-lactamase-like protein LACTB | | Descriptor: | Serine beta-lactamase-like protein LACTB, mitochondrial | | Authors: | Zhang, M.H, Yang, M.J. | | Deposit date: | 2021-08-07 | | Release date: | 2022-02-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis for the catalytic activity of filamentous human serine beta-lactamase-like protein LACTB.
Structure, 30, 2022
|
|
7DL8
 
 | |
