6GIZ
 
 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - SUBSTRATE COMPLEX | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
6GJ2
 
 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - COMPLEX WITH INOSITOL HEXASULPHATE | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
6GJ9
 
 | |
6GIT
 
 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - PRODUCT COMPLEX | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.418 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
6GJA
 
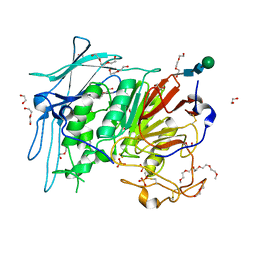 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - H229A MUTANT | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-16 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
1PNB
 
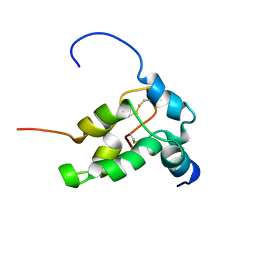 | | STRUCTURE OF NAPIN BNIB, NMR, 10 STRUCTURES | | Descriptor: | NAPIN BNIB | | Authors: | Rico, M, Bruix, M, Gonzalez, C, Monsalve, R, Rodriguez, R. | | Deposit date: | 1996-09-17 | | Release date: | 1997-09-17 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | 1H NMR assignment and global fold of napin BnIb, a representative 2S albumin seed protein.
Biochemistry, 35, 1996
|
|
1SM7
 
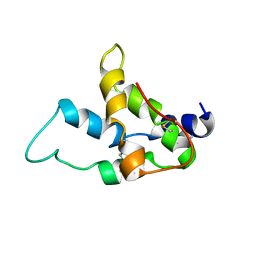 | | Solution structure of the recombinant pronapin precursor, BnIb. | | Descriptor: | recombinant Ib pronapin | | Authors: | Pantoja-Uceda, D, Palomares, O, Bruix, M, Villalba, M, Rodriguez, R, Rico, M, Santoro, J. | | Deposit date: | 2004-03-08 | | Release date: | 2005-02-01 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure and stability against digestion of rproBnIb, a recombinant 2S albumin from rapeseed: relationship to its allergenic properties.
Biochemistry, 43, 2004
|
|
1SS3
 
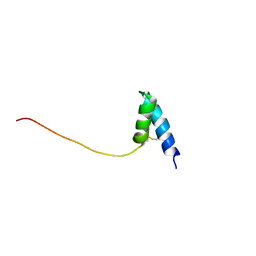 | | Solution structure of Ole e 6, an allergen from olive tree pollen | | Descriptor: | Pollen allergen Ole e 6 | | Authors: | Trevino, M.A, Garcia-Mayoral, M.F, Barral, P, Villalba, M, Santoro, J, Rico, M, Rodriguez, R, Bruix, M. | | Deposit date: | 2004-03-23 | | Release date: | 2004-08-03 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Ole e 6, a Major Allergen from Olive Tree Pollen.
J.Biol.Chem., 279, 2004
|
|
6FK2
 
 | |
2JON
 
 | | Solution structure of the C-terminal domain Ole e 9 | | Descriptor: | Beta-1,3-glucanase | | Authors: | Trevino, M.A, Palomares, O, Castrillo, I, Villalba, M, Rodriguez, R, Rico, M, Santoro, J, Bruix, M. | | Deposit date: | 2007-03-14 | | Release date: | 2008-01-29 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of Ole e 9, a major allergen of olive pollen
Protein Sci., 17, 2008
|
|
2MUA
 
 | |
2MUE
 
 | | Structure Immunogenicity and Protectivity Relationship for the 1585 Malarial Peptide and Its Substitution Analogues | | Descriptor: | Merozoite surface protein 1 | | Authors: | Espejo, F, Cubillos, M, Salazar, L, Guzman, F, Urquiza, M, Ocampo, M, Silva, Y, Rodriguez, R, Lioy, E, Patarroyo, M. | | Deposit date: | 2014-09-08 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Immunogenicity, and Protectivity Relationship for the 1585 Malarial Peptide and Its Substitution Analogues.
Angew.Chem.Int.Ed.Engl., 40, 2001
|
|
2MU8
 
 | | Residues belonging the n-terminal region derived of merozoite surface protein-2 of plasmodium falciparum | | Descriptor: | MSP-2 peptide | | Authors: | Cifuentes, G, Patarroyo, M.E, Urquiza, M, Ramirez, L.E, Reyes, C, Rodriguez, R. | | Deposit date: | 2014-09-04 | | Release date: | 2014-11-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Distorting malaria peptide backbone structure to enable fitting into MHC class II molecules renders modified peptides immunogenic and protective.
J.Med.Chem., 46, 2003
|
|
2MU7
 
 | | Shortening and modifying the 1513 MSP-1 peptide's alpha-helical region induces protection against malaria | | Descriptor: | 1513 MSP-1 peptide | | Authors: | Espejo, F, Bermudez, A, Torres, E, Urquiza, M, Rodriguez, R, Lopez, Y, Patarroyo, M. | | Deposit date: | 2014-09-04 | | Release date: | 2014-11-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Shortening and modifying the 1513 MSP-1 peptide's alpha-helical region induces protection against malaria.
Biochem.Biophys.Res.Commun., 315, 2004
|
|
6ZUC
 
 | | X-RAY CRYSTAL STRUCTURE OF THE CsPYL1-Lig1-HAB1 TERNARY COMPLEX | | Descriptor: | 1,4-dimethyl-2-oxidanylidene-~{N}-(phenylmethyl)quinoline-6-sulfonamide, CHLORIDE ION, CSPYL1, ... | | Authors: | Albert, A, Infantes, L, Benavente, J.L. | | Deposit date: | 2020-07-22 | | Release date: | 2021-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-guided engineering of a receptor-agonist pair for inducible activation of the ABA adaptive response to drought.
Sci Adv, 9, 2023
|
|
8AY3
 
 | |
8AY6
 
 | | X-RAY CRYSTAL STRUCTURE OF THE CsPYL1(V112L, T135L,F137I, T153I, V168A)-SB-HAB1 TERNARY COMPLEX | | Descriptor: | 1,4-dimethyl-2-oxidanylidene-~{N}-(phenylmethyl)quinoline-6-sulfonamide, Abscisic acid receptor PYL1, CHLORIDE ION, ... | | Authors: | Infantes, L, Albert, A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure-guided engineering of a receptor-agonist pair for inducible activation of the ABA adaptive response to drought.
Sci Adv, 9, 2023
|
|
8AY8
 
 | | X-RAY CRYSTAL STRUCTURE OF THE CsPYL1(V112L, T135L,F137I, T153I, V168A)-iSB9-HAB1 TERNARY COMPLEX | | Descriptor: | Abscisic acid receptor PYL1, CHLORIDE ION, GLYCEROL, ... | | Authors: | Infantes, L, Albert, A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-guided engineering of a receptor-agonist pair for inducible activation of the ABA adaptive response to drought.
Sci Adv, 9, 2023
|
|
8AYA
 
 | |
8AY9
 
 | | X-RAY CRYSTAL STRUCTURE OF THE CsPYL1(V112L, T135L,F137I, T153I, V168A)-ABA-HAB1 TERNARY COMPLEX | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL1, CHLORIDE ION, ... | | Authors: | Infantes, L, Albert, A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | Structure-guided engineering of a receptor-agonist pair for inducible activation of the ABA adaptive response to drought.
Sci Adv, 9, 2023
|
|
8AY7
 
 | | X-RAY CRYSTAL STRUCTURE OF THE CsPYL1(V112L, T135L,F137I, T153I, V168A)-iSB7-HAB1 TERNARY COMPLEX | | Descriptor: | Abscisic acid receptor PYL1, CHLORIDE ION, GLYCEROL, ... | | Authors: | Infantes, L, Albert, A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.131 Å) | | Cite: | Structure-guided engineering of a receptor-agonist pair for inducible activation of the ABA adaptive response to drought.
Sci Adv, 9, 2023
|
|
2OPO
 
 | | Crystal structure of the calcium-binding pollen allergen Che a 3 | | Descriptor: | CALCIUM ION, Polcalcin Che a 3, SULFATE ION | | Authors: | Verdino, P, Keller, W. | | Deposit date: | 2007-01-29 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Three-dimensional structure of the cross-reactive pollen allergen Che a 3: visualizing cross-reactivity on the molecular surfaces of weed, grass, and tree pollen allergens.
J.Immunol., 180, 2008
|
|
8OXE
 
 | | Inositol 1,3,4-trisphosphate 5/6-kinase 1 from Solanum tuberosum (StITPK1) in complex with ADP/Mg2+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Inositol-tetrakisphosphate 1-kinase, MAGNESIUM ION | | Authors: | Faba-Rodriguez, R, Li, A.W.H, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2023-05-02 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal Structure and Enzymology of Solanum tuberosum Inositol Tris/Tetrakisphosphate Kinase 1 ( St ITPK1).
Biochemistry, 63, 2024
|
|
6BFX
 
 | | BACE crystal structure with hydroxy pyrrolidine inhibitor | | Descriptor: | Beta-secretase 1, GLYCEROL, N-{(1S,2S)-3-(3,5-difluorophenyl)-1-[(3R,5S,6R)-6-(2,2-dimethylpropoxy)-5-methylmorpholin-3-yl]-1-hydroxypropan-2-yl}acetamide | | Authors: | Timm, D.E. | | Deposit date: | 2017-10-27 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Optimization of Hydroxyethylamine Transition State Isosteres as Aspartic Protease Inhibitors by Exploiting Conformational Preferences.
J. Med. Chem., 60, 2017
|
|
6BFE
 
 | | BACE crystal structure with hydroxy pyrrolidine inhibitor | | Descriptor: | Beta-secretase 1, GLYCEROL, N-[(1R,2S)-1-[(2R,4R)-4-(cyclohexylmethoxy)pyrrolidin-2-yl]-3-(3,5-difluorophenyl)-1-hydroxypropan-2-yl]acetamide | | Authors: | Timm, D.E. | | Deposit date: | 2017-10-26 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Optimization of Hydroxyethylamine Transition State Isosteres as Aspartic Protease Inhibitors by Exploiting Conformational Preferences.
J. Med. Chem., 60, 2017
|
|
