2R9G
 
 | | Crystal structure of the C-terminal fragment of AAA ATPase from Enterococcus faecium | | Descriptor: | AAA ATPase, central region, ACETATE ION, ... | | Authors: | Ramagopal, U.A, Patskovsky, Y, Bonanno, J.B, Shi, W, Toro, R, Meyer, A.J, Rutter, M, Wu, B, Groshong, C, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-09-12 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of the C-Terminal Domain of AAA ATPase from Enterococcus faecium.
To be Published
|
|
3DBY
 
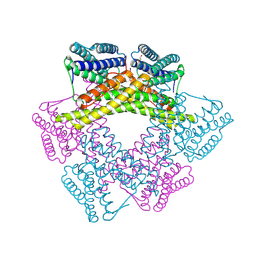 | | Crystal structure of uncharacterized protein from Bacillus cereus G9241 (CSAP Target) | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, uncharacterized protein | | Authors: | Ramagopal, U.A, Bonanno, J.B, Ozyurt, S, Freeman, J, Wasserman, S, Hu, S, Groshong, C, Rodgers, L, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-02 | | Release date: | 2008-07-29 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of uncharacterized protein from Bacillus cereus G9241
To be published
|
|
5TRU
 
 | | Structure of the first-in-class checkpoint inhibitor Ipilimumab bound to human CTLA-4 | | Descriptor: | Cytotoxic T-lymphocyte protein 4, Ipilimumab Fab heavy chain, Ipilimumab Fab light chain | | Authors: | Ramagopal, U.A, Liu, W, Garrett-Thomson, S.C, Yan, Q, Srinivasan, M, Wong, S.C, Bell, A, Mankikar, S, Rangan, V.S, Deshpande, S, Bonanno, J.B, Korman, A.J, Almo, S.C. | | Deposit date: | 2016-10-27 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for cancer immunotherapy by the first-in-class checkpoint inhibitor ipilimumab.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1RZN
 
 | |
1TWY
 
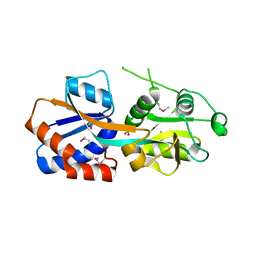 | | Crystal structure of an ABC-type phosphate transport receptor from Vibrio cholerae | | Descriptor: | ABC transporter, periplasmic substrate-binding protein, MAGNESIUM ION, ... | | Authors: | Ramagopal, U.A, Patskovsky, Y, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-01 | | Release date: | 2004-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of hypothetical ABC-type phosphate transporter
To be Published
|
|
4ETO
 
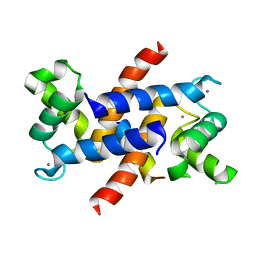 | | Structure of S100A4 in complex with non-muscle myosin-IIA peptide | | Descriptor: | CALCIUM ION, Myosin-9, Protein S100-A4 | | Authors: | Ramagopal, U.A, Dulyaninova, N.G, Kumar, P.R, Almo, S.C, Bresnick, A.R, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-04-24 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure of S100A4 with bound peptide P
To be Published
|
|
2A1V
 
 | |
2FIQ
 
 | |
1OAD
 
 | | Glucose isomerase from Streptomyces rubiginosus in P21212 crystal form | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Ramagopal, U.A, Dauter, M, Dauter, Z. | | Deposit date: | 2003-01-08 | | Release date: | 2003-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Sad Manganese in Two Crystal Forms of Glucose Isomerase
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1RKQ
 
 | |
3LYB
 
 | |
3LYP
 
 | |
3M0W
 
 | | Structure of S100A4 with PCP | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-chloro-10-[3-(4-methylpiperazin-1-yl)propyl]-10H-phenothiazine, CALCIUM ION, ... | | Authors: | Ramagopal, U.A, Dulyaninova, N.G, Almo, S.C, Bresnick, A.R. | | Deposit date: | 2010-03-03 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of S100A4 with PCP
To be published
|
|
3MDN
 
 | |
3ME5
 
 | | Crystal structure of putative dna cytosine methylase from shigella flexneri 2a str. 2457T | | Descriptor: | Cytosine-specific methyltransferase | | Authors: | Ramagopal, U.A, Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-21 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of putative dna cytosine methylase from shigella flexneri 2a str. 2457T
To be Published
|
|
3MDK
 
 | |
4HSZ
 
 | | Structure of truncated (delta8C) S100A4 | | Descriptor: | CALCIUM ION, Protein S100-A4 | | Authors: | Ramagopal, U.A, Dulyaninova, N.G, Kumar, P.R, Almo, S.C, Bresnick, A.R, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-10-31 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of truncated (delta8C) S100A4
To be Published
|
|
4HZA
 
 | |
3QLD
 
 | |
3RQ3
 
 | | Structure of T-cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT) in hexagonal crystal form | | Descriptor: | CHLORIDE ION, T cell immunoreceptor with Ig and ITIM domains | | Authors: | Ramagopal, U.A, Rubinstein, R, Guo, H, Samanta, D, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2011-04-27 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of T-cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT) in hexagonal crystal form
To be published
|
|
3S4T
 
 | | Crystal structure of putative amidohydrolase-2 (EFI-target 500288)from Polaromonas sp. JS666 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, Amidohydrolase 2, ... | | Authors: | Ramagopal, U.A, Toro, R, Girlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-05-20 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of putative amidohydrolase-2 (EFI-target 500288)from Polaromonas sp. JS666
To be published
|
|
3S5S
 
 | |
3N1U
 
 | | Structure of putative HAD superfamily (subfamily III A) hydrolase from Legionella pneumophila | | Descriptor: | CALCIUM ION, Hydrolase, HAD superfamily, ... | | Authors: | Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-17 | | Release date: | 2010-07-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of putative HAD superfamily (subfamily III A) hydrolase from Legionella pneumophila
To be published
|
|
3PR8
 
 | | Structure of Glutathione S-transferase(PP0183) from Pseudomonas putida in complex with GSH | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLUTATHIONE, Glutathione S-transferase family protein | | Authors: | Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-11-29 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Glutathione S-transferase(PP0183) from Pseudomonas putida in complex with GSH
To be Published
|
|
3Q0H
 
 | | Structure of T-cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT) | | Descriptor: | T cell immunoreceptor with Ig and ITIM domains | | Authors: | Ramagopal, U.A, Guo, H, Samanta, D, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-12-15 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of T-cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT)
To be published
|
|
