5ID3
 
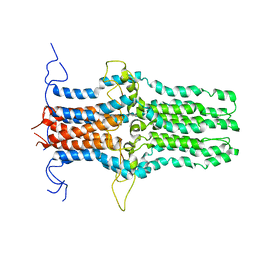 | | Solution structure of the pore-forming region of C. elegans Mitochondrial Calcium Uniporter (MCU) | | Descriptor: | Mitochondrial Calcium Uniporter | | Authors: | Oxenoid, K, Dong, Y, Cao, C, Cui, T, Sancak, Y, Markhard, A.L, Grabarek, Z, Kong, L, Liu, Z, Ouyang, B, Cong, Y, Mootha, V.K, Chou, J.J, Membrane Protein Structures by Solution NMR (MPSbyNMR) | | Deposit date: | 2016-02-23 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Architecture of the mitochondrial calcium uniporter.
Nature, 533, 2016
|
|
8OM2
 
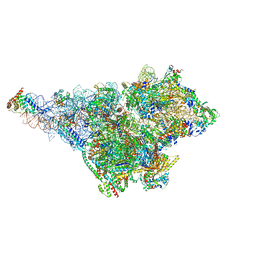 | | Small subunit of yeast mitochondrial ribosome in complex with METTL17/Rsm22. | | Descriptor: | 15S mitochondrial rRNA, 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial, ... | | Authors: | Itoh, Y, Chicherin, I, Kamenski, P, Amunts, A. | | Deposit date: | 2023-03-31 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | METTL17 is an Fe-S cluster checkpoint for mitochondrial translation.
Mol.Cell, 84, 2024
|
|
8OM4
 
 | | Small subunit of yeast mitochondrial ribosome. | | Descriptor: | 15S mitochondrial rRNA, 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial, ... | | Authors: | Itoh, Y, Chicherin, I, Kamenski, P, Amunts, A. | | Deposit date: | 2023-03-31 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | METTL17 is an Fe-S cluster checkpoint for mitochondrial translation.
Mol.Cell, 84, 2024
|
|
8OM3
 
 | | Small subunit of yeast mitochondrial ribosome in complex with IF3/Aim23. | | Descriptor: | 15S mitochondrial rRNA, 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial, ... | | Authors: | Itoh, Y, Chicherin, I, Kamenski, P, Amunts, A. | | Deposit date: | 2023-03-31 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | METTL17 is an Fe-S cluster checkpoint for mitochondrial translation.
Mol.Cell, 84, 2024
|
|
5ER0
 
 | |
6EAZ
 
 | |
6OXD
 
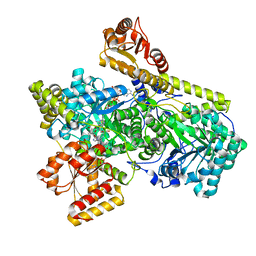 | | Structure of Mycobacterium tuberculosis methylmalonyl-CoA mutase with adenosyl cobalamin | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, Itaconyl coenzyme A, ... | | Authors: | Purchal, M, Ruetz, M, Banerjee, R, Koutmos, M. | | Deposit date: | 2019-05-13 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Itaconyl-CoA forms a stable biradical in methylmalonyl-CoA mutase and derails its activity and repair.
Science, 366, 2019
|
|
6OXC
 
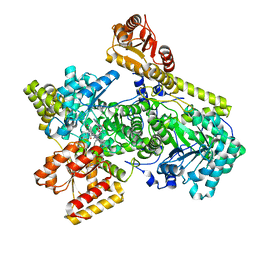 | | Structure of Mycobacterium tuberculosis methylmalonyl-CoA mutase with adenosyl cobalamin | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, Methylmalonyl-CoA mutase large subunit, ... | | Authors: | Purchal, M, Ruetz, M, Banerjee, R, Koutmos, M. | | Deposit date: | 2019-05-13 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Itaconyl-CoA forms a stable biradical in methylmalonyl-CoA mutase and derails its activity and repair.
Science, 366, 2019
|
|
6U08
 
 | |
6XJX
 
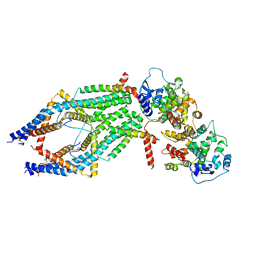 | | MCU holocomplex in Low-calcium blocking state | | Descriptor: | Calcium uniporter protein, mitochondrial, Calcium uptake protein 1, ... | | Authors: | Wang, Y, Jiang, Y. | | Deposit date: | 2020-06-24 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural insights into the Ca 2+ -dependent gating of the human mitochondrial calcium uniporter.
Elife, 9, 2020
|
|
6XJV
 
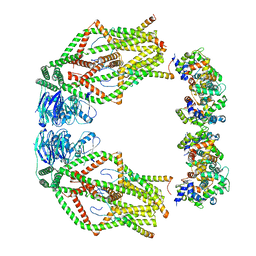 | | MCU holocomplex in High-calcium state | | Descriptor: | Calcium uniporter protein, mitochondrial, Calcium uptake protein 1, ... | | Authors: | Wang, Y, Jiang, Y. | | Deposit date: | 2020-06-24 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Structural insights into the Ca 2+ -dependent gating of the human mitochondrial calcium uniporter.
Elife, 9, 2020
|
|
5VXC
 
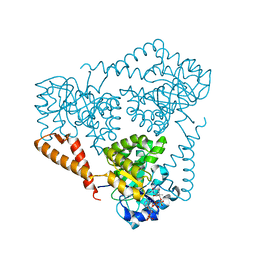 | |
5VOH
 
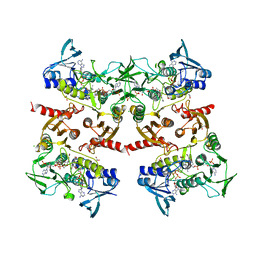 | | Crystal structure of engineered water-forming NADPH oxidase (TPNOX) bound to NADPH. The G159A, D177A, A178R, M179S, P184R mutant of LbNOX. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cracan, V, Grabarek, Z. | | Deposit date: | 2017-05-02 | | Release date: | 2017-08-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | A genetically encoded tool for manipulation of NADP(+)/NADPH in living cells.
Nat. Chem. Biol., 13, 2017
|
|
5VN0
 
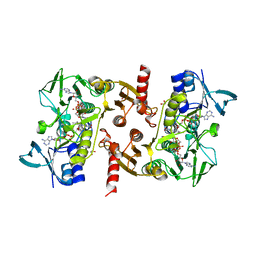 | | Water-forming NADH oxidase from Lactobacillus brevis (LbNOX) bound to NADH. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cracan, V, Grabarek, Z. | | Deposit date: | 2017-04-28 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | A genetically encoded tool for manipulation of NADP(+)/NADPH in living cells.
Nat. Chem. Biol., 13, 2017
|
|
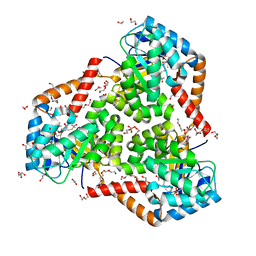
5VXO
 
 | |
5VXS
 
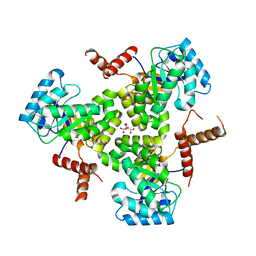 | | Crystal Structure Analysis of human CLYBL in apo form | | Descriptor: | CITRIC ACID, Citrate lyase subunit beta-like protein, mitochondrial | | Authors: | Shen, H. | | Deposit date: | 2017-05-24 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | The Human Knockout Gene CLYBL Connects Itaconate to Vitamin B12.
Cell, 171, 2017
|
|
6D80
 
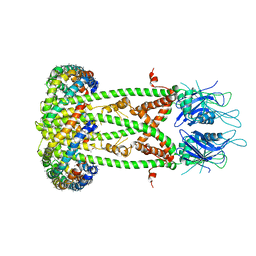 | | Cryo-EM structure of the mitochondrial calcium uniporter from N. fischeri bound to saposin | | Descriptor: | CALCIUM ION, Mitochondrial calcium uniporter, Saposin A | | Authors: | Nguyen, N.X, Armache, J.-P, Cheng, Y, Bai, X.C. | | Deposit date: | 2018-04-25 | | Release date: | 2018-07-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Cryo-EM structure of a fungal mitochondrial calcium uniporter.
Nature, 559, 2018
|
|
6D7W
 
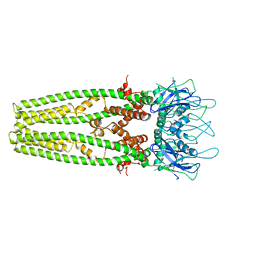 | | Cryo-EM structure of the mitochondrial calcium uniporter from N. fischeri at 3.8 Angstrom resolution | | Descriptor: | CALCIUM ION, Mitochondrial calcium uniporter | | Authors: | Nguyen, N.X, Armache, J.-P, Cheng, Y, Bai, X.C. | | Deposit date: | 2018-04-25 | | Release date: | 2018-07-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of a fungal mitochondrial calcium uniporter.
Nature, 559, 2018
|
|
7THF
 
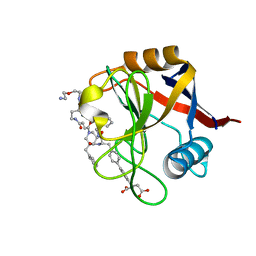 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B53 | | Descriptor: | 3-(4'-{[(4S,7S,11R,13E,19S)-19-{[2-(2-aminoethoxy)ethyl]carbamoyl}-7-benzyl-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosin-4-yl]methyl}[1,1'-biphenyl]-4-yl)pentanedioic acid, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
7TH1
 
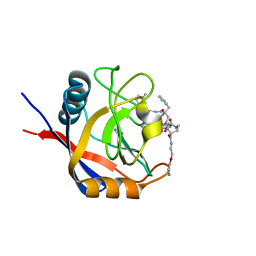 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B3 | | Descriptor: | (4S,7S,11R,13E,19S)-N-[2-(2-aminoethoxy)ethyl]-7-benzyl-4-[(3-methylphenyl)methyl]-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosine-19-carboxamide, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
7TGV
 
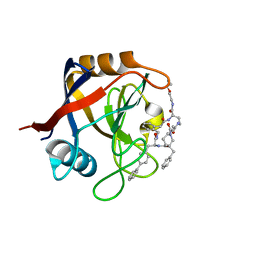 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B2 | | Descriptor: | (4S,7S,11R,13E,19S)-N-[2-(2-aminoethoxy)ethyl]-7-benzyl-4-[([1,1'-biphenyl]-4-yl)methyl]-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosine-19-carboxamide, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-09 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
7TGS
 
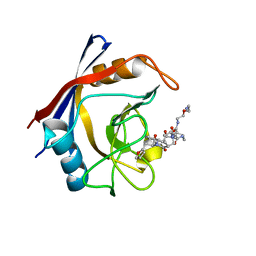 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor JOMBt | | Descriptor: | (4S,7R,11S,13E,19S)-N-[2-(2-aminoethoxy)ethyl]-4-[(furan-2-yl)methyl]-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosine-19-carboxamide, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-09 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
7TH7
 
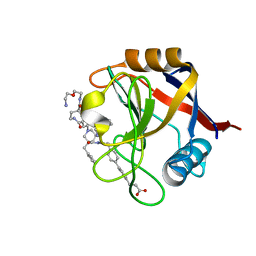 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B23 | | Descriptor: | 3-(4'-{[(4S,7S,11R,19S)-19-{[2-(2-aminoethoxy)ethyl]carbamoyl}-7-benzyl-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,13,14,15,16,17,18,19,20,21,22-icosahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosin-4-yl]methyl}[1,1'-biphenyl]-4-yl)propanoic acid, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
7TGU
 
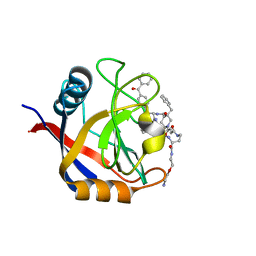 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B1 | | Descriptor: | (4S,7S,11R,13E,19S)-N-[2-(2-aminoethoxy)ethyl]-4-[(4-benzoylphenyl)methyl]-7-benzyl-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosine-19-carboxamide, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-09 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
7THC
 
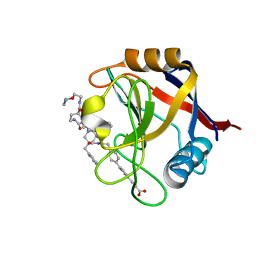 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B25 | | Descriptor: | 3-(4'-{[(4S,7S,11R,13E,19S)-19-{[2-(2-aminoethoxy)ethyl]carbamoyl}-7-benzyl-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosin-4-yl]methyl}[1,1'-biphenyl]-4-yl)prop-2-ynoic acid, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
