3F5C
 
 | | Structure of Dax-1:LRH-1 complex | | Descriptor: | Nuclear receptor subfamily 0 group B member 1, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Fletterick, R.J, Sablin, E.P. | | Deposit date: | 2008-11-03 | | Release date: | 2008-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of corepressor Dax-1 bound to its target nuclear receptor LRH-1.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1ANC
 
 | |
1AND
 
 | |
1ANB
 
 | |
1ANE
 
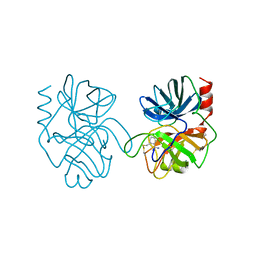 | | ANIONIC TRYPSIN WILD TYPE | | Descriptor: | ANIONIC TRYPSIN, BENZAMIDINE | | Authors: | Fletterick, R.J, Mcgrath, M.E. | | Deposit date: | 1994-12-21 | | Release date: | 1997-04-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of an engineered, metal-actuated switch in trypsin.
Biochemistry, 32, 1993
|
|
1BSX
 
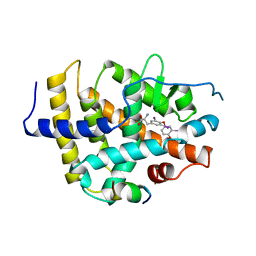 | | STRUCTURE AND SPECIFICITY OF NUCLEAR RECEPTOR-COACTIVATOR INTERACTIONS | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, PROTEIN (GRIP1), PROTEIN (THYROID HORMONE RECEPTOR BETA) | | Authors: | Wagner, R.L, Darimont, B.D, Apriletti, J.W, Stallcup, M.R, Kushner, P.J, Baxter, J.D, Fletterick, R.J, Yamamoto, K.R. | | Deposit date: | 1998-08-31 | | Release date: | 1999-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure and specificity of nuclear receptor-coactivator interactions.
Genes Dev., 12, 1998
|
|
3LVH
 
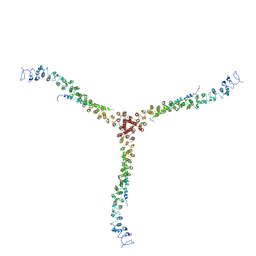 | | Crystal structure of a clathrin heavy chain and clathrin light chain complex | | Descriptor: | Clathrin heavy chain 1, Clathrin light chain B | | Authors: | Wilbur, J.D, Hwang, P.K, Ybe, J.A, Lane, M, Sellers, B.D, Jacobson, M.P, Fletterick, R.J, Brodsky, F.M. | | Deposit date: | 2010-02-20 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (9 Å) | | Cite: | Conformation switching of clathrin light chain regulates clathrin lattice assembly.
Dev.Cell, 18, 2010
|
|
3CNZ
 
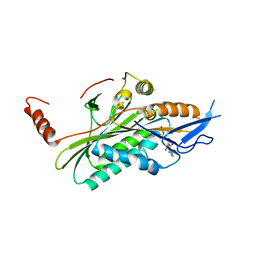 | | Structural dynamics of the microtubule binding and regulatory elements in the kinesin-like calmodulin binding protein | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin heavy chain-like protein, MAGNESIUM ION | | Authors: | Vinogradova, M.V, Malanina, G.G, Reddy, V, Reddy, A.S.N, Fletterick, R.J. | | Deposit date: | 2008-03-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural dynamics of the microtubule binding and regulatory elements in the kinesin-like calmodulin binding protein.
J.Struct.Biol., 163, 2008
|
|
1SLW
 
 | |
1SLX
 
 | |
1SLU
 
 | |
1SLV
 
 | |
2Z4J
 
 | |
3COB
 
 | | Structural Dynamics of the Microtubule binding and regulatory elements in the Kinesin-like Calmodulin binding protein | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin heavy chain-like protein, MAGNESIUM ION | | Authors: | Vinogradova, M.V, Malanina, G.G, Reddy, V, Reddy, A.S.N, Fletterick, R.J. | | Deposit date: | 2008-03-27 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural dynamics of the microtubule binding and regulatory elements in the kinesin-like calmodulin binding protein.
J.Struct.Biol., 163, 2008
|
|
1SDM
 
 | | Crystal structure of kinesin-like calmodulin binding protein | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, kinesin heavy chain-like protein | | Authors: | Vinogradova, M.V, Reddy, V.S, Reddy, A.S, Sablin, E.P, Fletterick, R.J. | | Deposit date: | 2004-02-13 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of kinesin regulated by Ca(2+)-calmodulin.
J.Biol.Chem., 279, 2004
|
|
1CR9
 
 | | CRYSTAL STRUCTURE OF THE ANTI-PRION FAB 3F4 | | Descriptor: | FAB ANTIBODY HEAVY CHAIN, FAB ANTIBODY LIGHT CHAIN | | Authors: | Kanyo, Z.F, Pan, K.M, Williamson, R.A, Burton, D.R, Prusiner, S.B, Fletterick, R.J, Cohen, F.E. | | Deposit date: | 1999-08-14 | | Release date: | 2000-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antibody binding defines a structure for an epitope that participates in the PrPC-->PrPSc conformational change.
J.Mol.Biol., 293, 1999
|
|
1CU4
 
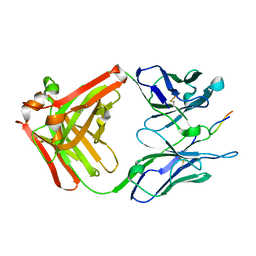 | | CRYSTAL STRUCTURE OF THE ANTI-PRION FAB 3F4 IN COMPLEX WITH ITS PEPTIDE EPITOPE | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, RECOGNITION PEPTIDE | | Authors: | Kanyo, Z.F, Pan, K.M, Williamson, R.A, Burton, D.R, Prusiner, S.B, Fletterick, R.J, Cohen, F.E. | | Deposit date: | 1999-08-20 | | Release date: | 2000-04-17 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Antibody binding defines a structure for an epitope that participates in the PrPC-->PrPSc conformational change.
J.Mol.Biol., 293, 1999
|
|
1YGP
 
 | | PHOSPHORYLATED FORM OF YEAST GLYCOGEN PHOSPHORYLASE WITH PHOSPHATE BOUND IN THE ACTIVE SITE. | | Descriptor: | PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, YEAST GLYCOGEN PHOSPHORYLASE | | Authors: | Lin, K, Rath, V.L, Dai, S.C, Fletterick, R.J, Hwang, P.K. | | Deposit date: | 1996-05-30 | | Release date: | 1996-12-23 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A protein phosphorylation switch at the conserved allosteric site in GP.
Science, 273, 1996
|
|
6DDA
 
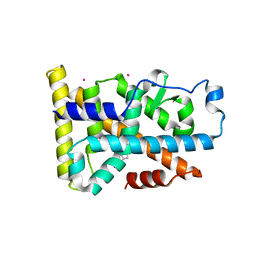 | | Nurr1 Covalently Modified by a Dopamine Metabolite | | Descriptor: | 5-hydroxy-1,2-dihydro-6H-indol-6-one, BROMIDE ION, Nuclear receptor subfamily 4 group A member 2, ... | | Authors: | Bruning, J.M, Wang, Y, Otrabella, F, Boxue, T, Liu, H, Bhattacharya, P, Guo, S, Holton, J.M, Fletterick, R.J, Jacobson, M.P, England, P.M. | | Deposit date: | 2018-05-09 | | Release date: | 2019-03-20 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Covalent Modification and Regulation of the Nuclear Receptor Nurr1 by a Dopamine Metabolite.
Cell Chem Biol, 26, 2019
|
|
3LVG
 
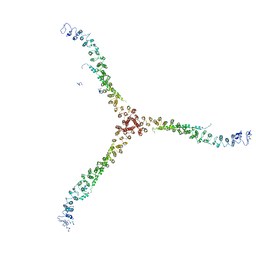 | | Crystal structure of a clathrin heavy chain and clathrin light chain complex | | Descriptor: | Clathrin heavy chain 1, Clathrin light chain B | | Authors: | Wilbur, J.D, Hwang, P.K, Ybe, J.A, Lane, M, Sellers, B.D, Jacobson, M.P, Fletterick, R.J, Brodsky, F.M. | | Deposit date: | 2010-02-20 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (7.94 Å) | | Cite: | Conformation switching of clathrin light chain regulates clathrin lattice assembly.
Dev.Cell, 18, 2010
|
|
2AIM
 
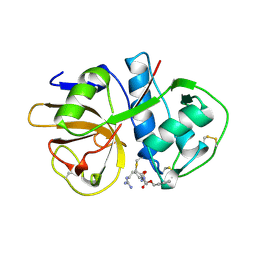 | |
3F7D
 
 | | SF-1 LBD bound by phosphatidylcholine | | Descriptor: | (2S)-2-{[(1R)-1-hydroxyhexadecyl]oxy}-3-{[(1R)-1-hydroxyoctadecyl]oxy}propyl 2-(trimethylammonio)ethyl phosphate, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, nuclear receptor SF-1 | | Authors: | Sablin, E.P, Fletterick, R.J. | | Deposit date: | 2008-11-07 | | Release date: | 2008-12-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of SF-1 bound by different phospholipids: evidence for regulatory ligands.
Mol.Endocrinol., 23, 2009
|
|
3I00
 
 | | Crystal Structure of the huntingtin interacting protein 1 coiled coil domain | | Descriptor: | Huntingtin-interacting protein 1 | | Authors: | Wilbur, J.D, Hwang, P.K, Brodsky, F.M, Fletterick, R.J. | | Deposit date: | 2009-06-24 | | Release date: | 2010-02-23 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Accommodation of structural rearrangements in the huntingtin-interacting protein 1 coiled-coil domain.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1A0Q
 
 | | 29G11 COMPLEXED WITH PHENYL [1-(1-N-SUCCINYLAMINO)PENTYL] PHOSPHONATE | | Descriptor: | 29G11 FAB (HEAVY CHAIN), 29G11 FAB (LIGHT CHAIN), PHENYL[1-(N-SUCCINYLAMINO)PENTYL]PHOSPHONATE, ... | | Authors: | Buchbinder, J.L, Stephenson, R.C, Scanlan, T.S, Fletterick, R.J. | | Deposit date: | 1997-12-05 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A comparison of the crystallographic structures of two catalytic antibodies with esterase activity.
J.Mol.Biol., 282, 1998
|
|
1AZZ
 
 | | FIDDLER CRAB COLLAGENASE COMPLEXED TO ECOTIN | | Descriptor: | COLLAGENASE, ECOTIN | | Authors: | Perona, J.J, Fletterick, R.J. | | Deposit date: | 1997-11-24 | | Release date: | 1998-02-25 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an ecotin-collagenase complex suggests a model for recognition and cleavage of the collagen triple helix.
Biochemistry, 36, 1997
|
|
