6LFU
 
 | | Poa1p F152A mutant in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose 1''-phosphate phosphatase | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.123 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
6LFT
 
 | | Poa1p S30A mutant in complex with ADP-ribose | | Descriptor: | ACETATE ION, ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose 1''-phosphate phosphatase | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
6LFS
 
 | | Poa1p H23A mutant in complex with ADP-ribose | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose 1''-phosphate phosphatase | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
6LFQ
 
 | | Crystal structure of Poa1p in apo form | | Descriptor: | ADP-ribose 1''-phosphate phosphatase, GLYCEROL | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
6LFR
 
 | | Poa1p in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose 1''-phosphate phosphatase | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
7F2V
 
 | | Urate oxidase from Thermobispora bispora in apo form | | Descriptor: | DIMETHYL SULFOXIDE, SULFATE ION, Uricase | | Authors: | Chiu, Y.C, Hsu, T.S, Huang, C.Y, Hsu, C.H. | | Deposit date: | 2021-06-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and biochemical insights into a hyperthermostable urate oxidase from Thermobispora bispora for hyperuricemia and gout therapy.
Int.J.Biol.Macromol., 188, 2021
|
|
7F2W
 
 | | TbUox in complex with uric acid | | Descriptor: | URIC ACID, Uricase | | Authors: | Chiu, Y.C, Hsu, T.S, Huang, C.Y, Hsu, C.H. | | Deposit date: | 2021-06-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural and biochemical insights into a hyperthermostable urate oxidase from Thermobispora bispora for hyperuricemia and gout therapy.
Int.J.Biol.Macromol., 188, 2021
|
|
2CWV
 
 | | Product schiff-base intermediate of copper amine oxidase from arthrobacter globiformis | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Chiu, Y.C, Okajima, T, Murakawa, T, Uchida, M, Taki, M, Hirota, S, Kim, M, Yamaguchi, H, Kawano, Y, Kamiya, N, Kuroda, S, Hayashi, H, Yamamoto, Y, Tanizawa, K. | | Deposit date: | 2005-06-26 | | Release date: | 2006-05-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Kinetic and Structural Studies on the Catalytic Role of the Aspartic Acid Residue Conserved in Copper Amine Oxidase(,)
Biochemistry, 45, 2006
|
|
2CWU
 
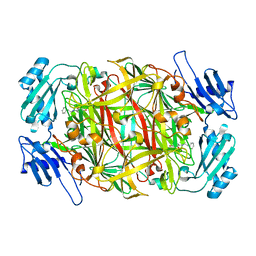 | | Substrate schiff-base intermediate of copper amine oxidase from arthrobacter globiformis | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Chiu, Y.C, Okajima, T, Murakawa, T, Uchida, M, Taki, M, Hirota, S, Kim, M, Yamaguchi, H, Kawano, Y, Kamiya, N, Kuroda, S, Hayashi, H, Yamamoto, Y, Tanizawa, K. | | Deposit date: | 2005-06-26 | | Release date: | 2006-05-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Kinetic and Structural Studies on the Catalytic Role of the Aspartic Acid Residue Conserved in Copper Amine Oxidase(,)
Biochemistry, 45, 2006
|
|
2CWT
 
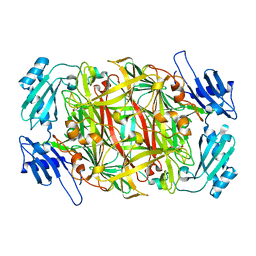 | | Catalytic base deletion in copper amine oxidase from arthrobacter globiformis | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Chiu, Y.C, Okajima, T, Murakawa, T, Uchida, M, Taki, M, Hirota, S, Kim, M, Yamaguchi, H, Kawano, Y, Kamiya, N, Kuroda, S, Hayashi, H, Yamamoto, Y, Tanizawa, K. | | Deposit date: | 2005-06-26 | | Release date: | 2006-05-02 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Kinetic and Structural Studies on the Catalytic Role of the Aspartic Acid Residue Conserved in Copper Amine Oxidase(,)
Biochemistry, 45, 2006
|
|
6DD2
 
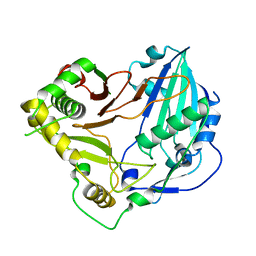 | | Crystal structure of Selaginella moellendorffii HCT | | Descriptor: | Probable hydroxycinnamoyl transferase | | Authors: | Levsh, O, Chiang, Y.C, Lam, C.K, Wang, Y, Weng, J.K. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9056 Å) | | Cite: | Structural and dynamic basis of substrate permissiveness in hydroxycinnamoyltransferase (HCT).
PLoS Comput. Biol., 14, 2018
|
|
8JBQ
 
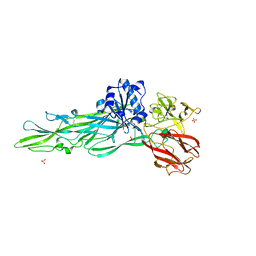 | |
6OH1
 
 | | IgA1 Protease G5 domain structure | | Descriptor: | Immunoglobulin A1 protease | | Authors: | Eisenmesser, E.Z, Chi, Y.C, Paukovich, N, Redzic, J.S, Rahkola, J.T, Janoff, E.N. | | Deposit date: | 2019-04-04 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Streptococcus pneumoniae G5 domains bind different ligands.
Protein Sci., 28, 2019
|
|
5KJS
 
 | | Crystal Structure of Arabidopsis thaliana HCT | | Descriptor: | Shikimate O-hydroxycinnamoyltransferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
5KJV
 
 | | Crystal structure of Coleus blumei HCT | | Descriptor: | Hydroxycinnamoyl transferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
5KJT
 
 | | Crystal structure of Arabidopsis thaliana HCT in complex with p-coumaroyl-CoA | | Descriptor: | Shikimate O-hydroxycinnamoyltransferase, p-coumaroyl-CoA | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
5KJW
 
 | | Crystal structure of Coleus blumei HCT in complex with 3-hydroxyacetophenone | | Descriptor: | 1-(3-hydroxyphenyl)ethanone, Hydroxycinnamoyl transferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
5KJU
 
 | | Crystal structure of Arabidopsis thaliana HCT in complex with p-coumaroylshikimate | | Descriptor: | (3~{R},4~{S},5~{R})-3-[(~{E})-3-(4-hydroxyphenyl)prop-2-enoyl]oxy-4,5-bis(oxidanyl)cyclohexene-1-carboxylic acid, Shikimate O-hydroxycinnamoyltransferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
6DXF
 
 | |
6DXB
 
 | |
6DXA
 
 | |
6DX9
 
 | |
6DXC
 
 | |
6DXE
 
 | |
6DX8
 
 | |
