2NMQ
 
 | |
4CO6
 
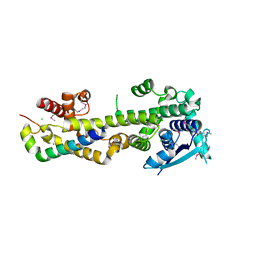 | | Crystal structure of the Nipah virus RNA free nucleoprotein- phosphoprotein complex | | Descriptor: | BROMIDE ION, CHLORIDE ION, NUCLEOPROTEIN, ... | | Authors: | Yabukarksi, F, Lawrence, P, Tarbouriech, N, Bourhis, J.M, Jensen, M.R, Ruigrok, R.W.H, Blackledge, M, Volchkov, V, Jamin, M. | | Deposit date: | 2014-01-27 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Structure of Nipah Virus Unassembled Nucleoprotein in Complex with its Viral Chaperone.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4UX9
 
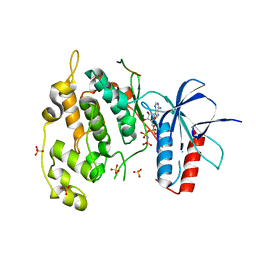 | | Crystal structure of JNK1 bound to a MKK7 docking motif | | Descriptor: | DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 7, MITOGEN-ACTIVATED PROTEIN KINASE 8, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Kragelj, J, Palencia, A, Nanao, M.H, Maurin, D, Bouvignies, G, Blackledge, M, Ringkjobing-Jensen, M. | | Deposit date: | 2014-08-20 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure and Dynamics of the Mkk7-Jnk Signaling Complex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2P7C
 
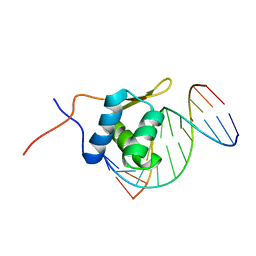 | | Solution structure of the bacillus licheniformis BlaI monomeric form in complex with the blaP half-operator. | | Descriptor: | Penicillinase repressor, Strand 1 of Twelve base-pair DNA, Strand 2 of Twelve base-pair DNA | | Authors: | Boudet, J, Duval, V, Van Melckebeke, H, Blackledge, M, Amoroso, A, Joris, B, Simorre, J.-P. | | Deposit date: | 2007-03-20 | | Release date: | 2007-06-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformational and thermodynamic changes of the repressor/DNA operator complex upon monomerization shed new light on regulation mechanisms of bacterial resistance against beta-lactam antibiotics.
Nucleic Acids Res., 35, 2007
|
|
1YKG
 
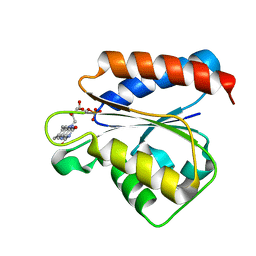 | | Solution structure of the flavodoxin-like domain from the Escherichia coli sulfite reductase | | Descriptor: | FLAVIN MONONUCLEOTIDE, Sulfite reductase [NADPH] flavoprotein alpha-component | | Authors: | Sibille, N, Blackledge, M, Brutscher, B, Coves, J, Bersch, B. | | Deposit date: | 2005-01-18 | | Release date: | 2005-07-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Sulfite Reductase Flavodoxin-like Domain from Escherichia coli
Biochemistry, 44, 2005
|
|
7PKU
 
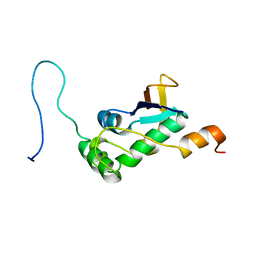 | | Structure of SARS-CoV-2 nucleoprotein in dynamic complex with its viral partner nsp3a | | Descriptor: | 3C-like proteinase, Nucleoprotein | | Authors: | Bessa, L.M, Guseva, S, Camacho-Zarco, A.R, Salvi, N, Blackledge, M. | | Deposit date: | 2021-08-26 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The intrinsically disordered SARS-CoV-2 nucleoprotein in dynamic complex with its viral partner nsp3a.
Sci Adv, 8, 2022
|
|
8CLR
 
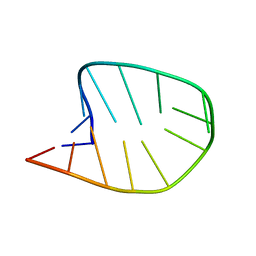 | | Integrated NMR/MD structure determination of a dynamic and thermodynamically stable CUUG RNA tetraloop | | Descriptor: | RNA hairpin with CUUG tetraloop | | Authors: | Oxenfarth, A, Kuemmerer, F, Bottaro, S, Schnieders, R, Pinter, G, Jonker, H.R.A, Fuertig, B, Richter, C, Blackledge, M, Lindorff-Larsen, K, Schwalbe, H. | | Deposit date: | 2023-02-17 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Integrated NMR/Molecular Dynamics Determination of the Ensemble Conformation of a Thermodynamically Stable CUUG RNA Tetraloop.
J.Am.Chem.Soc., 145, 2023
|
|
6H5Q
 
 | | Cryo-EM structure of in vitro assembled Measles virus N into nucleocapsid-like particles (NCLPs) bound to polyA RNA hexamers. | | Descriptor: | Nucleocapsid, RNA (5'-R(*AP*AP*AP*AP*AP*A)-3') | | Authors: | Desfosses, A, Milles, S, Ringkjobing Jensen, M, Guseva, S, Colletier, J, Maurin, D, Schoehn, G, Gutsche, I, Ruigrok, R, Blackledge, M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Assembly and cryo-EM structures of RNA-specific measles virus nucleocapsids provide mechanistic insight into paramyxoviral replication.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2PLP
 
 | |
6H5S
 
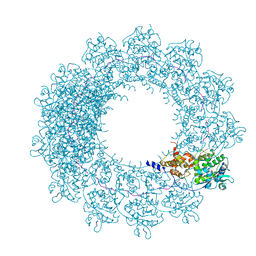 | | Cryo-EM map of in vitro assembled Measles virus N into nucleocapsid-like particles (NCLPs) bound to viral genomic 5-prime RNA hexamers. | | Descriptor: | Nucleocapsid, RNA (5'-R(*AP*CP*CP*AP*GP*A)-3') | | Authors: | Desfosses, A, Milles, S, Ringkjobing Jensen, M, Guseva, S, Colletier, J.P, Maurin, D, Schoehn, G, Gutsche, I, Ruigrok, R, Blackledge, M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Assembly and cryo-EM structures of RNA-specific measles virus nucleocapsids provide mechanistic insight into paramyxoviral replication.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1C2N
 
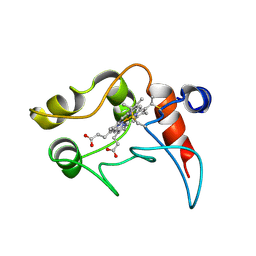 | | CYTOCHROME C2, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C2, HEME C | | Authors: | Cordier, F, Caffrey, M.S, Brutscher, B, Cusanovich, M.A, Marion, D, Blackledge, M. | | Deposit date: | 1998-04-27 | | Release date: | 1999-03-23 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure, rotational diffusion anisotropy and local backbone dynamics of Rhodobacter capsulatus cytochrome c2.
J.Mol.Biol., 281, 1998
|
|
6TRI
 
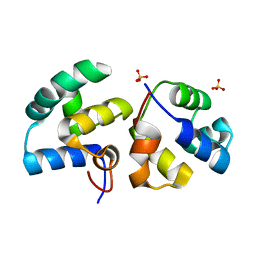 | | CI-MOR repressor-antirepressor complex of the temperate bacteriophage TP901-1 from Lactococcus lactis | | Descriptor: | CI, MOR, SULFATE ION | | Authors: | Rasmussen, K.K, Blackledge, M, Herrmann, T, Jensen, M.R, Lo Leggio, L. | | Deposit date: | 2019-12-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.277 Å) | | Cite: | Revealing the mechanism of repressor inactivation during switching of a temperate bacteriophage.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6TO6
 
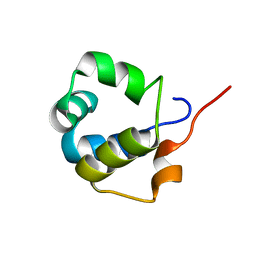 | | Solution structure of the modulator of repression (MOR) of the temperate bacteriophage TP901-1 from Lactococcus lactis | | Descriptor: | MOR | | Authors: | Rasmussen, K.K, Blackledge, M, Herrmann, T, Lo Leggio, L, Jensen, M.R. | | Deposit date: | 2019-12-11 | | Release date: | 2020-08-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Revealing the mechanism of repressor inactivation during switching of a temperate bacteriophage.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1R4G
 
 | | Solution structure of the Sendai virus protein X C-subdomain | | Descriptor: | RNA polymerase alpha subunit | | Authors: | Blanchard, L, Tarbouriech, N, Blackledge, M, Timmins, P, Burmeister, W.P, Ruigrok, R.W, Marion, D. | | Deposit date: | 2003-10-06 | | Release date: | 2004-03-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of the nucleocapsid-binding domain of the Sendai virus phosphoprotein in solution
Virology, 319, 2004
|
|
1IMT
 
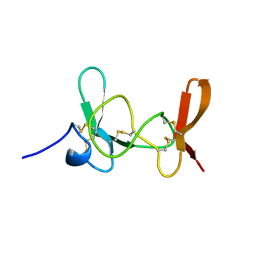 | | MAMBA INTESTINAL TOXIN 1, NMR, 39 STRUCTURES | | Descriptor: | INTESTINAL TOXIN 1 | | Authors: | Boisbouvier, J, Albrand, J.-P, Blackledge, M, Jaquinod, M, Schweitz, H, Lazdunski, M, Marion, D. | | Deposit date: | 1998-04-14 | | Release date: | 1999-04-20 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | A structural homologue of colipase in black mamba venom revealed by NMR floating disulphide bridge analysis.
J.Mol.Biol., 283, 1998
|
|
1EKY
 
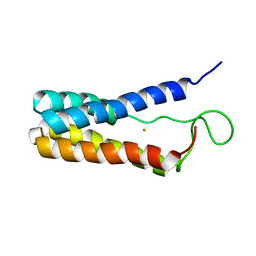 | |
3ZDO
 
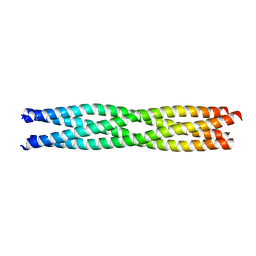 | | Tetramerization domain of Measles virus phosphoprotein | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, PHOSPHOPROTEIN | | Authors: | Communie, G, Crepin, T, Jensen, M.R, Blackledge, M, Ruigrok, R.W.H. | | Deposit date: | 2012-11-29 | | Release date: | 2013-04-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure of the Tetramerization Domain of Measles Virus Phosphoprotein.
J.Virol., 87, 2013
|
|
4A56
 
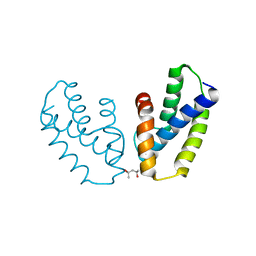 | | Crystal structure of the type 2 secretion system pilotin from Klebsiella Oxytoca | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, PULLULANASE SECRETION PROTEIN PULS | | Authors: | Tosi, T, Nickerson, N.N, Mollica, L, RingkjobingJensen, M, Blackledge, M, Baron, B, England, P, Pugsley, A.P, Dessen, A. | | Deposit date: | 2011-10-24 | | Release date: | 2011-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Pilotin-Secretin Recognition in the Type II Secretion System of Klebsiella Oxytoca.
Mol.Microbiol, 82, 2011
|
|
1FCT
 
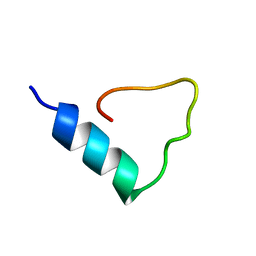 | |
1I5V
 
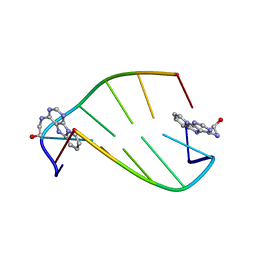 | | SOLUTION STRUCTURE OF 2-(PYRIDO[1,2-E]PURIN-4-YL)AMINO-ETHANOL INTERCALATED IN THE DNA DUPLEX D(CGATCG)2 | | Descriptor: | 2-(PYRIDO[1,2-E]PURIN-4-YL)AMINO-ETHANOL, 5'-D(*CP*GP*AP*TP*CP*G)-3' | | Authors: | Favier, A, Blackledge, M, Simorre, J.P, Marion, D, Debousy, J.C. | | Deposit date: | 2001-03-01 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of 2-(pyrido[1,2-e]purin-4-yl)amino-ethanol intercalated in the DNA duplex d(CGATCG)2.
Biochemistry, 40, 2001
|
|
1K1C
 
 | | Solution Structure of Crh, the Bacillus subtilis Catabolite Repression HPr | | Descriptor: | catabolite repression HPr-like protein | | Authors: | Favier, A, Brutscher, B, Blackledge, M, Galinier, A, Deutscher, J, Penin, F, Marion, D. | | Deposit date: | 2001-09-25 | | Release date: | 2001-10-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of Crh, the Bacillus subtilis catabolite repression HPr.
J.Mol.Biol., 317, 2002
|
|
2KRO
 
 | |
2MCN
 
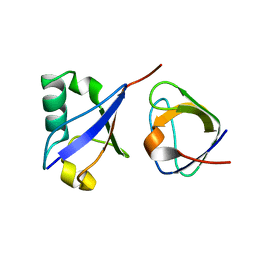 | | Distinct ubiquitin binding modes exhibited by SH3 domains: molecular determinants and functional implications | | Descriptor: | CD2-associated protein, Ubiquitin | | Authors: | Ortega-Roldan, J, Salmon, L, Azuaga, A, Blackledge, M, Van Nuland, N. | | Deposit date: | 2013-08-22 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Distinct ubiquitin binding modes exhibited by SH3 domains: molecular determinants and functional implications.
Plos One, 8, 2013
|
|
2K47
 
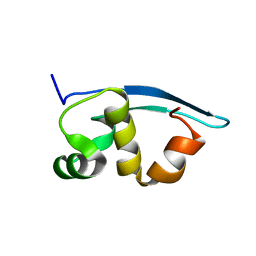 | | Solution structure of the C-terminal N-RNA binding domain of the Vesicular Stomatitis Virus Phosphoprotein | | Descriptor: | Phosphoprotein | | Authors: | Ribeiro, E.A, Favier, A, Gerard, F.C, Leyrat, C, Brutscher, B, Blondel, D, Ruigrok, R.W, Blackledge, M, Jamin, M. | | Deposit date: | 2008-05-28 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-Terminal Nucleoprotein-RNA Binding Domain of the Vesicular Stomatitis Virus Phosphoprotein.
J.Mol.Biol., 2008
|
|
2LZ6
 
 | | Distinct ubiquitin binding modes exhibited by sh3 domains: molecular determinants and functional implications | | Descriptor: | CD2-associated protein, Ubiquitin | | Authors: | Ortega-Roldan, J, Azuaga, A, Blackledge, M, Van Nuland, N. | | Deposit date: | 2012-09-24 | | Release date: | 2013-10-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Distinct Ubiquitin Binding Modes Exhibited by SH3 Domains: Molecular Determinants and Functional Implications.
Plos One, 8, 2013
|
|
