8IZJ
 
 | |
5GHU
 
 | | Crystal structure of YadV chaperone at 1.63 Angstrom | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Pandey, N.K, Bhavesh, N.S. | | Deposit date: | 2016-06-20 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of the usher chaperone YadV reveals a monomer with the proline lock in closed conformation suggestive of an intermediate state.
Febs Lett., 2020
|
|
6LE1
 
 | | Structure of RRM2 domain of DND1 protein | | Descriptor: | Dead end protein homolog 1, GLYCEROL, SULFATE ION | | Authors: | Kumari, P, Bhavesh, N.S. | | Deposit date: | 2019-11-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human DND1-RRM2 forms a non-canonical domain swapped dimer.
Protein Sci., 30, 2021
|
|
6M75
 
 | | C-Myc DNA binding protein complex | | Descriptor: | DNA (5'-D(*TP*CP*TP*TP*AP*TP*T)-3'), RNA-binding motif, single-stranded-interacting protein 1, ... | | Authors: | Aggarwal, P, Bhavesh, N.S. | | Deposit date: | 2020-03-17 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Hinge like domain motion facilitates human RBMS1 protein binding to proto-oncogene c-myc promoter.
Nucleic Acids Res., 49, 2021
|
|
4EYV
 
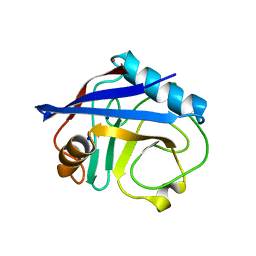 | | Crystal structure of Cyclophilin A like protein from Piriformospora indica | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Peptidyl-prolyl cis-trans isomerase, ... | | Authors: | Bhatt, H, Pal, R.K, Tuteja, N, Bhavesh, N.S. | | Deposit date: | 2012-05-02 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of RNA-interacting Cyclophilin A-like protein from Piriformospora indica that provides salinity-stress tolerance in plants
Sci Rep, 3, 2013
|
|
7C36
 
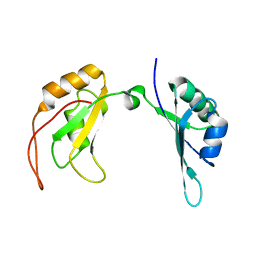 | | c-Myc DNA binding protein structure | | Descriptor: | RNA-binding motif, single-stranded-interacting protein 1 | | Authors: | Aggarwal, P, Bhavesh, N.S. | | Deposit date: | 2020-05-11 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Hinge like domain motion facilitates human RBMS1 protein binding to proto-oncogene c-myc promoter.
Nucleic Acids Res., 49, 2021
|
|
1EVO
 
 | | NMR OBSERVATION OF A NOVEL C-TETRAD | | Descriptor: | DNA (5'-D(*TP*GP*GP*GP*CP*GP*GP*T)-3') | | Authors: | Patel, P.K, Bhavesh, N.S, Hosur, R.V. | | Deposit date: | 2000-04-20 | | Release date: | 2000-05-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR observation of a novel C-tetrad in the structure of the SV40 repeat sequence GGGCGG.
Biochem.Biophys.Res.Commun., 270, 2000
|
|
7Y94
 
 | |
7XI2
 
 | |
7WEZ
 
 | |
1L1K
 
 | |
4TLQ
 
 | |
1LV1
 
 | | Crystal Structure Analysis of the non-active site mutant of tethered HIV-1 protease to 2.1A resolution | | Descriptor: | HIV-1 protease | | Authors: | Kumar, M, Kannan, K.K, Hosur, M.V, Bhavesh, N.S, Chatterjee, A, Mittal, R, Hosur, R.V. | | Deposit date: | 2002-05-24 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effects of remote mutation on the autolysis of HIV-1 PR: X-ray and NMR investigations.
Biochem.Biophys.Res.Commun., 294, 2002
|
|
2MMY
 
 | |
5IHF
 
 | |
5IO8
 
 | | Salmonella Typhimurium VirG-like (STV) protein at 2.19 Angstrom resolution solved by Iodine SAD. | | Descriptor: | IODIDE ION, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Pandey, N.K, Ray, S, Suar, M, Bhavesh, N.S. | | Deposit date: | 2016-03-08 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | Salmonella Typhimurium VirG-like (STV) protein at at 2.19 Angstrom resolution by SAD.
To Be Published
|
|
4LJM
 
 | |
4LMZ
 
 | |
5ZVN
 
 | | Structure of [beta Glc-T9,K7]indolicidin, a glycosylated analogue of indolicidin | | Descriptor: | beta-D-glucopyranose, glycosylated analogue of Indolicidin | | Authors: | Dwivedi, R, Aggarwal, P, Kaur, K.J, Bhavesh, N.S. | | Deposit date: | 2018-05-11 | | Release date: | 2019-06-19 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Design of therapeutically improved analogue of the antimicrobial peptide, indolicidin, using a glycosylation strategy.
Amino Acids, 51, 2019
|
|
5ZVF
 
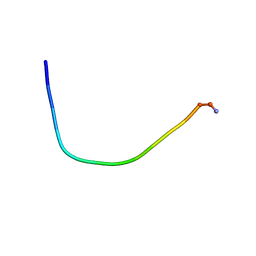 | | Structure of [T9,K7]indolicidin, a non glycosylated analogue of indolicidin | | Descriptor: | non glycosylated analogue of Indolicidin | | Authors: | Dwivedi, R, Aggarwal, P, Kaur, K.J, Bhavesh, N.S. | | Deposit date: | 2018-05-10 | | Release date: | 2019-06-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Design of therapeutically improved analogue of the antimicrobial peptide, indolicidin, using a glycosylation strategy.
Amino Acids, 51, 2019
|
|
2MY7
 
 | |
2MYF
 
 | |
2MY8
 
 | |
2N3L
 
 | |
2N7C
 
 | |
