5UZF
 
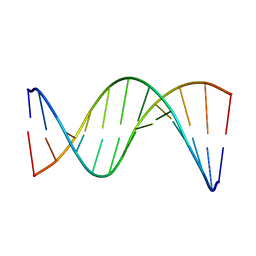 | | Insights into Watson-Crick/Hoogsteen Breathing Dynamics and Damage Repair from the Solution Structure and Dynamic Ensemble of DNA Duplexes containing m1A - A6-DNA structure | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*TP*TP*TP*TP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*AP*AP*AP*AP*AP*TP*CP*G)-3') | | Authors: | Sathyamoorthy, B, Shi, H, Xue, Y, Al-Hashimi, H.M. | | Deposit date: | 2017-02-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into Watson-Crick/Hoogsteen breathing dynamics and damage repair from the solution structure and dynamic ensemble of DNA duplexes containing m1A.
Nucleic Acids Res., 45, 2017
|
|
5UZD
 
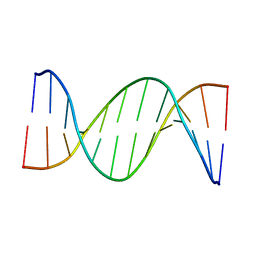 | | Insights into Watson-Crick/Hoogsteen Breathing Dynamics and Damage Repair from the Solution Structure and Dynamic Ensemble of DNA Duplexes containing m1A - A2-DNA structure | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*GP*AP*TP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*AP*TP*CP*GP*AP*TP*GP*C)-3') | | Authors: | Sathyamoorthy, B, Shi, H, Xue, Y, Al-Hashimi, H.M. | | Deposit date: | 2017-02-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into Watson-Crick/Hoogsteen breathing dynamics and damage repair from the solution structure and dynamic ensemble of DNA duplexes containing m1A.
Nucleic Acids Res., 45, 2017
|
|
5UZI
 
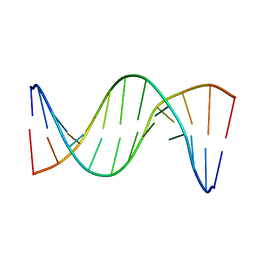 | | Insights into Watson-Crick/Hoogsteen Breathing Dynamics and Damage Repair from the Solution Structure and Dynamic Ensemble of DNA Duplexes containing m1A - A6-DNAm1A16 structure | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*TP*TP*TP*TP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*(M1A)P*AP*AP*AP*AP*AP*TP*CP*G)-3') | | Authors: | Sathyamoorthy, B, Shi, H, Xue, Y, Al-Hashimi, H.M. | | Deposit date: | 2017-02-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into Watson-Crick/Hoogsteen breathing dynamics and damage repair from the solution structure and dynamic ensemble of DNA duplexes containing m1A.
Nucleic Acids Res., 45, 2017
|
|
8U3M
 
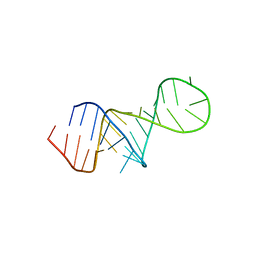 | | The FARFAR-MD-NMR ensemble of an HIV-1 TAR excited state | | Descriptor: | The excited state of HIV-1 transactivation response element (31-MER) | | Authors: | Geng, A, Ganser, L, Roy, R, Shi, H, Pratihar, S, Case, D.A, Al-Hashimi, H.M. | | Deposit date: | 2023-09-07 | | Release date: | 2023-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An RNA excited conformational state at atomic resolution.
Nat Commun, 14, 2023
|
|
8THV
 
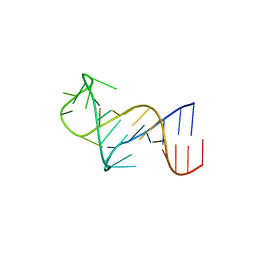 | | FARFAR-NMR ensemble of HIV-1 TAR with apical loop capturing ground and excited conformational states | | Descriptor: | RNA (29-MER) | | Authors: | Roy, R, Geng, A, Shi, H, Merriman, D.K, Dethoff, E.A, Salmon, L, Al-Hashimi, H.M. | | Deposit date: | 2023-07-18 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Kinetic Resolution of the Atomic 3D Structures Formed by Ground and Excited Conformational States in an RNA Dynamic Ensemble.
J.Am.Chem.Soc., 145, 2023
|
|
7JU1
 
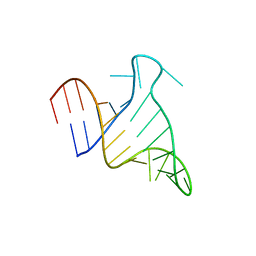 | | The FARFAR-NMR Ensemble of 29-mer HIV-1 Trans-activation Response Element RNA (N=20) | | Descriptor: | RNA (29-MER) | | Authors: | Shi, H, Rangadurai, A, Roy, R, Yesselman, J.D, Al-Hashimi, H.M. | | Deposit date: | 2020-08-18 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rapid and accurate determination of atomistic RNA dynamic ensemble models using NMR and structure prediction
Nat Commun, 11, 2020
|
|
1FH1
 
 | |
6UEP
 
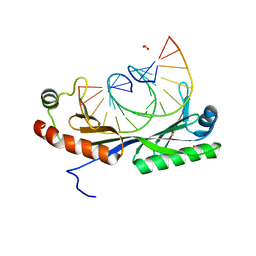 | |
6UER
 
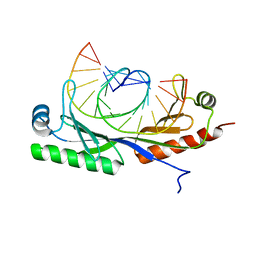 | |
6UEQ
 
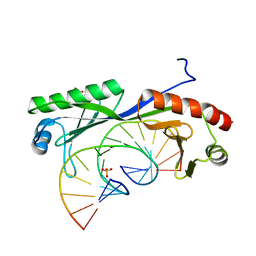 | | Structure of TBP bound to C-C mismatch containing TATA site | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*AP*AP*CP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*TP*TP*TP*TP*AP*TP*AP*GP*C)-3'), SULFATE ION, ... | | Authors: | Schumacher, M.A, Al-Hashimi, H. | | Deposit date: | 2019-09-22 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | DNA mismatches reveal conformational penalties in protein-DNA recognition.
Nature, 587, 2020
|
|
6NJQ
 
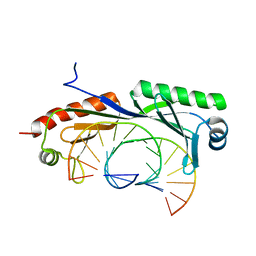 | | Structure of TBP-Hoogsteen containing DNA complex | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*AP*CP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*GP*TP*TP*TP*AP*TP*AP*GP*C)-3'), TATA-box-binding protein 1 | | Authors: | Schumacher, M.A, Stelling, A. | | Deposit date: | 2019-01-04 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Infrared Spectroscopic Observation of a G-C+Hoogsteen Base Pair in the DNA:TATA-Box Binding Protein Complex Under Solution Conditions.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5DDQ
 
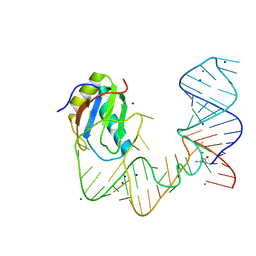 | | L-glutamine riboswitch bound with L-glutamine soaked with Mn2+ | | Descriptor: | GLUTAMINE, L-glutamine riboswitch RNA (61-MER), MAGNESIUM ION, ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
5DDR
 
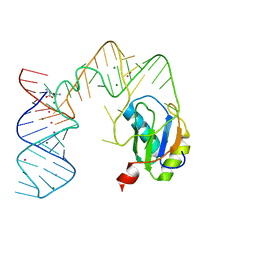 | | L-glutamine riboswitch bound with L-glutamine soaked with Cs+ | | Descriptor: | CESIUM ION, GLUTAMINE, L-glutamine riboswitch RNA (61-MER), ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
6E4N
 
 | |
6E4O
 
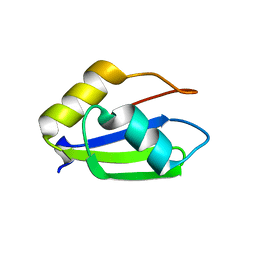 | | Structure of apo T. brucei RRM: P4(1)2(1)2 form | | Descriptor: | RNA-binding protein, putative | | Authors: | Schumacher, M.A. | | Deposit date: | 2018-07-18 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The RRM of the kRNA-editing protein TbRGG2 uses multiple surfaces to bind and remodel RNA.
Nucleic Acids Res., 47, 2019
|
|
6E4P
 
 | | Structure of the T. brucei RRM domain in complex with RNA | | Descriptor: | RNA (5'-R(P*UP*UP*UP*U)-3'), RNA-binding protein, putative | | Authors: | Schumacher, M.A. | | Deposit date: | 2018-07-18 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | The RRM of the kRNA-editing protein TbRGG2 uses multiple surfaces to bind and remodel RNA.
Nucleic Acids Res., 47, 2019
|
|
4J8O
 
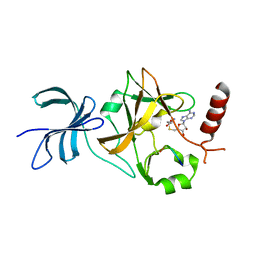 | | SET7/9 in complex with TAF10K189A peptide and AdoHcy | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, Transcription initiation factor TFIID subunit 10 | | Authors: | Horowitz, S, Trievel, R.C. | | Deposit date: | 2013-02-14 | | Release date: | 2014-01-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Conservation and functional importance of carbon-oxygen hydrogen bonding in AdoMet-dependent methyltransferases.
J.Am.Chem.Soc., 135, 2013
|
|
4J7I
 
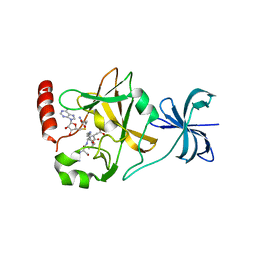 | | SET7/9Y335F in complex with TAF10 peptide and AdoHcy | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, Transcription initiation factor TFIID subunit 10 | | Authors: | Horowitz, S, Trievel, R.C. | | Deposit date: | 2013-02-13 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Conservation and functional importance of carbon-oxygen hydrogen bonding in AdoMet-dependent methyltransferases.
J.Am.Chem.Soc., 135, 2013
|
|
4J83
 
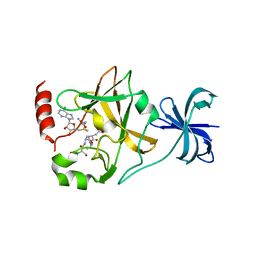 | | SET7/9 in complex with TAF10K189A peptide and AdoMet | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYLMETHIONINE, Transcription initiation factor TFIID subunit 10 | | Authors: | Horowitz, S, Nimtz, J.S, Trievel, R.C. | | Deposit date: | 2013-02-14 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conservation and functional importance of carbon-oxygen hydrogen bonding in AdoMet-dependent methyltransferases.
J.Am.Chem.Soc., 135, 2013
|
|
4J7F
 
 | |
5DDP
 
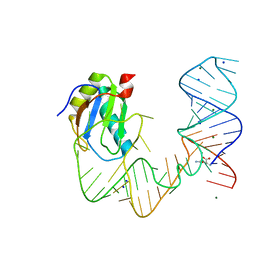 | | L-glutamine riboswitch bound with L-glutamine | | Descriptor: | GLUTAMINE, MAGNESIUM ION, RNA (61-MER), ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
5DDO
 
 | |
6M81
 
 | | Crystal structure of TylM1 Y14F bound to SAH and dTDP-phenol | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phenoxy)phosphoryl]oxy}phosphoryl]thymidine, S-ADENOSYL-L-HOMOCYSTEINE, dTDP-3-amino-3,6-dideoxy-alpha-D-glucopyranose N,N-dimethyltransferase | | Authors: | Fick, R.J, McDole, B.G, Trievel, R.C. | | Deposit date: | 2018-08-21 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | Structural and Functional Characterization of Sulfonium Carbon-Oxygen Hydrogen Bonding in the Deoxyamino Sugar Methyltransferase TylM1.
Biochemistry, 58, 2019
|
|
6M82
 
 | | Crystal structure of TylM1 Y14paF bound to SAH and dTDP-phenol | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phenoxy)phosphoryl]oxy}phosphoryl]thymidine, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Fick, R.J, McDole, B.G, Trievel, R.C. | | Deposit date: | 2018-08-21 | | Release date: | 2019-03-13 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.3971 Å) | | Cite: | Structural and Functional Characterization of Sulfonium Carbon-Oxygen Hydrogen Bonding in the Deoxyamino Sugar Methyltransferase TylM1.
Biochemistry, 58, 2019
|
|
6M83
 
 | | Crystal structure of TylM1 S120A bound to SAH and dTDP-phenol | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phenoxy)phosphoryl]oxy}phosphoryl]thymidine, CHLORIDE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Fick, R.J, McDole, B.G, Trievel, R.C. | | Deposit date: | 2018-08-21 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3685 Å) | | Cite: | Structural and Functional Characterization of Sulfonium Carbon-Oxygen Hydrogen Bonding in the Deoxyamino Sugar Methyltransferase TylM1.
Biochemistry, 58, 2019
|
|
