1J9L
 
 | | CRYSTAL STRUCTURE OF SURE PROTEIN FROM T.MARITIMA IN COMPLEX WITH VANADATE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, STATIONARY PHASE SURVIVAL PROTEIN, ... | | Authors: | Suh, S.W, Lee, J.Y, Kwak, J.E, Moon, J. | | Deposit date: | 2001-05-28 | | Release date: | 2001-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and functional analysis of the SurE protein identify a novel phosphatase family.
Nat.Struct.Biol., 8, 2001
|
|
1J9J
 
 | | CRYSTAL STRUCTURE ANALYSIS OF SURE PROTEIN FROM T.MARITIMA | | Descriptor: | MAGNESIUM ION, STATIONARY PHASE SURVIVAL PROTEIN, SULFATE ION | | Authors: | Suh, S.W, Lee, J.Y, Kwak, J.E, Moon, J. | | Deposit date: | 2001-05-27 | | Release date: | 2001-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and functional analysis of the SurE protein identify a novel phosphatase family.
Nat.Struct.Biol., 8, 2001
|
|
1J9K
 
 | | CRYSTAL STRUCTURE OF SURE PROTEIN FROM T.MARITIMA IN COMPLEX WITH TUNGSTATE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, STATIONARY PHASE SURVIVAL PROTEIN, ... | | Authors: | Suh, S.W, Lee, J.Y, Kwak, J.E, Moon, J. | | Deposit date: | 2001-05-27 | | Release date: | 2001-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and functional analysis of the SurE protein identify a novel phosphatase family.
Nat.Struct.Biol., 8, 2001
|
|
2GJL
 
 | | Crystal Structure of 2-nitropropane dioxygenase | | Descriptor: | FLAVIN MONONUCLEOTIDE, hypothetical protein PA1024 | | Authors: | Suh, S.W. | | Deposit date: | 2006-03-31 | | Release date: | 2006-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of 2-Nitropropane Dioxygenase Complexed with FMN and Substrate: identification of the catalytic base
J.Biol.Chem., 281, 2006
|
|
2GJN
 
 | |
2OWY
 
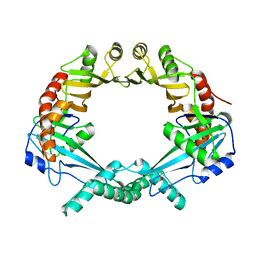 | |
1EW4
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI CYAY PROTEIN REVEALS A NOVEL FOLD FOR THE FRATAXIN FAMILY | | Descriptor: | CYAY PROTEIN | | Authors: | Suh, S.W, Cho, S, Lee, M.G, Yang, J.K, Lee, J.Y, Song, H.K. | | Deposit date: | 2000-04-22 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of Escherichia coli CyaY protein reveals a previously unidentified fold for the evolutionarily conserved frataxin family.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1RZL
 
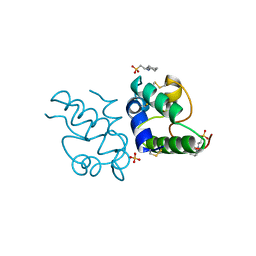 | | RICE NONSPECIFIC LIPID TRANSFER PROTEIN | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, NONSPECIFIC LIPID TRANSFER PROTEIN, SULFATE ION | | Authors: | Lee, J.Y, Min, K.S, Cha, H, Shin, D.H, Hwang, K.Y, Suh, S.W. | | Deposit date: | 1997-10-09 | | Release date: | 1998-12-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rice non-specific lipid transfer protein: the 1.6 A crystal structure in the unliganded state reveals a small hydrophobic cavity.
J.Mol.Biol., 276, 1998
|
|
3CNN
 
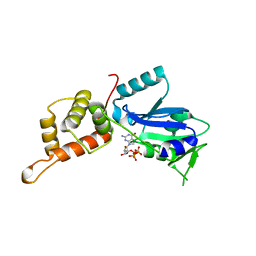 | | GTP-bound structure of TM YlqF | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Putative uncharacterized protein | | Authors: | Kim, D.J, Jang, J.Y, Yoon, H.-J, Suh, S.W. | | Deposit date: | 2008-03-26 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of YlqF, a circularly permuted GTPase: Implications for its GTPase activation in 50 S ribosomal subunit assembly
Proteins, 72, 2008
|
|
3DCM
 
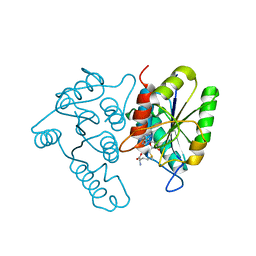 | | Crystal structure of the Thermotoga maritima SPOUT family RNA-methyltransferase protein Tm1570 in complex with S-adenosyl-L-methionine | | Descriptor: | S-ADENOSYLMETHIONINE, Uncharacterized protein TM_1570 | | Authors: | Kim, D.J, Kim, H.S, Lee, S.J, Suh, S.W. | | Deposit date: | 2008-06-04 | | Release date: | 2008-12-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Thermotoga maritima SPOUT superfamily RNA methyltransferase Tm1570 in complex with S-adenosyl-L-methionine
Proteins, 74, 2009
|
|
2RL2
 
 | |
2RL1
 
 | |
4KT6
 
 | | High-resolution crystal structure Streptococcus pyogenes beta-NAD+ glycohydrolase in complex with its endogenous inhibitor IFS reveals a water-rich interface | | Descriptor: | Nicotine adenine dinucleotide glycohydrolase, Putative uncharacterized protein | | Authors: | Yoon, J.Y, An, D.R, Yoon, H.-J, Kim, H.S, Lee, S.J, Im, H.N, Jang, J.Y, Suh, S.W. | | Deposit date: | 2013-05-20 | | Release date: | 2013-10-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | High-resolution crystal structure of Streptococcus pyogenes beta-NAD(+) glycohydrolase in complex with its endogenous inhibitor IFS reveals a highly water-rich interface
J.SYNCHROTRON RADIAT., 20, 2013
|
|
1CNS
 
 | |
1J2Z
 
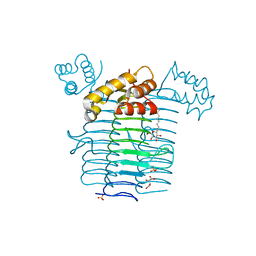 | | Crystal structure of UDP-N-acetylglucosamine acyltransferase | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, L(+)-TARTARIC ACID, SULFATE ION, ... | | Authors: | Lee, B.I, Suh, S.W. | | Deposit date: | 2003-01-15 | | Release date: | 2004-01-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of UDP-N-acetylglucosamine acyltransferase from Helicobacter pylori
Proteins, 53, 2003
|
|
1JXV
 
 | | Crystal Structure of Human Nucleoside Diphosphate Kinase A | | Descriptor: | Nucleoside Diphosphate Kinase A | | Authors: | Min, K, Song, H.K, Chang, C, Kim, S.Y, Lee, K.J, Suh, S.W. | | Deposit date: | 2001-09-10 | | Release date: | 2002-04-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human nucleoside diphosphate kinase A, a metastasis suppressor.
Proteins, 46, 2002
|
|
1MZL
 
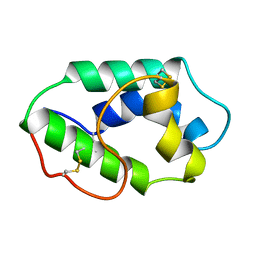 | | MAIZE NONSPECIFIC LIPID TRANSFER PROTEIN | | Descriptor: | MAIZE NONSPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Shin, D.H, Lee, J.Y, Hwang, K.Y, Kim, K.K, Suh, S.W. | | Deposit date: | 1995-01-26 | | Release date: | 1996-08-01 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structure of the non-specific lipid-transfer protein from maize seedlings.
Structure, 3, 1995
|
|
1MZM
 
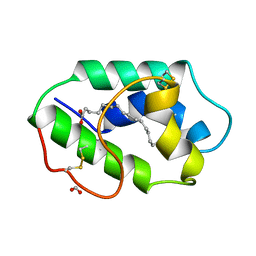 | |
5J1M
 
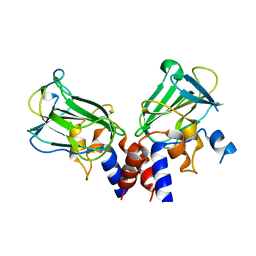 | | Crystal structure of Csd1-Csd2 dimer II | | Descriptor: | ToxR-activated gene (TagE), ZINC ION | | Authors: | An, D.R, Suh, S.W. | | Deposit date: | 2016-03-29 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of the Heterodimer Formation between Cell Shape-Determining Proteins Csd1 and Csd2 from Helicobacter pylori
Plos One, 11, 2016
|
|
5J1K
 
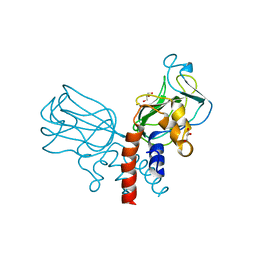 | | Crystal structure of Csd2-Csd2 dimer | | Descriptor: | GLYCEROL, ToxR-activated gene (TagE) | | Authors: | An, D.R, Suh, S.W. | | Deposit date: | 2016-03-29 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Basis of the Heterodimer Formation between Cell Shape-Determining Proteins Csd1 and Csd2 from Helicobacter pylori
Plos One, 11, 2016
|
|
5J1L
 
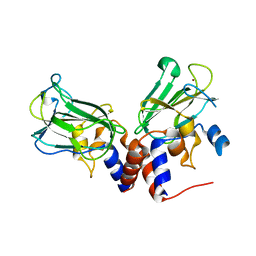 | | Crystal structure of Csd1-Csd2 dimer I | | Descriptor: | ToxR-activated gene (TagE), ZINC ION | | Authors: | An, D.R, Suh, S.W. | | Deposit date: | 2016-03-29 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural Basis of the Heterodimer Formation between Cell Shape-Determining Proteins Csd1 and Csd2 from Helicobacter pylori
Plos One, 11, 2016
|
|
1AVX
 
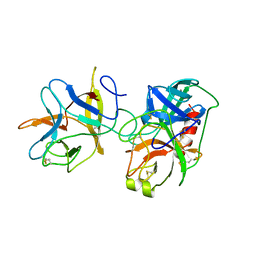 | | COMPLEX PORCINE PANCREATIC TRYPSIN/SOYBEAN TRYPSIN INHIBITOR, TETRAGONAL CRYSTAL FORM | | Descriptor: | CALCIUM ION, TRYPSIN, TRYPSIN INHIBITOR | | Authors: | Song, H.K, Suh, S.W. | | Deposit date: | 1997-09-21 | | Release date: | 1998-10-28 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kunitz-type soybean trypsin inhibitor revisited: refined structure of its complex with porcine trypsin reveals an insight into the interaction between a homologous inhibitor from Erythrina caffra and tissue-type plasminogen activator.
J.Mol.Biol., 275, 1998
|
|
6JN8
 
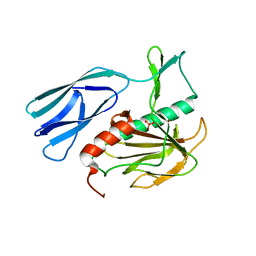 | | Structure of H216A mutant open form peptidoglycan peptidase | | Descriptor: | Peptidase M23, SULFATE ION, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
1AVU
 
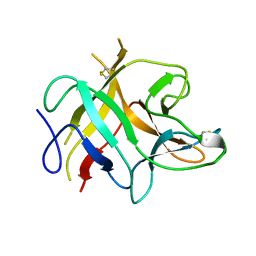 | | TRYPSIN INHIBITOR FROM SOYBEAN (STI) | | Descriptor: | TRYPSIN INHIBITOR | | Authors: | Song, H.K, Suh, S.W. | | Deposit date: | 1997-09-20 | | Release date: | 1998-10-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kunitz-type soybean trypsin inhibitor revisited: refined structure of its complex with porcine trypsin reveals an insight into the interaction between a homologous inhibitor from Erythrina caffra and tissue-type plasminogen activator.
J.Mol.Biol., 275, 1998
|
|
1AVW
 
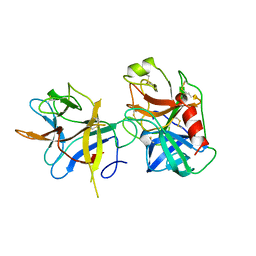 | | COMPLEX PORCINE PANCREATIC TRYPSIN/SOYBEAN TRYPSIN INHIBITOR, ORTHORHOMBIC CRYSTAL FORM | | Descriptor: | CALCIUM ION, TRYPSIN, TRYPSIN INHIBITOR | | Authors: | Song, H.K, Suh, S.W. | | Deposit date: | 1997-09-21 | | Release date: | 1998-10-28 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Kunitz-type soybean trypsin inhibitor revisited: refined structure of its complex with porcine trypsin reveals an insight into the interaction between a homologous inhibitor from Erythrina caffra and tissue-type plasminogen activator.
J.Mol.Biol., 275, 1998
|
|
