5V6P
 
 | | CryoEM structure of the ERAD-associated E3 ubiquitin-protein ligase HRD1 | | Descriptor: | ERAD-associated E3 ubiquitin-protein ligase HRD1 | | Authors: | Schoebel, S, Mi, W, Stein, A, Rapoport, T.A, Liao, M. | | Deposit date: | 2017-03-17 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3.
Nature, 548, 2017
|
|
3JZ9
 
 | | Crystal structure of the GEF domain of DrrA/SidM from Legionella pneumophila | | Descriptor: | Uncharacterized protein DrrA | | Authors: | Schoebel, S, Oesterlin, L.K, Blankenfeldt, W, Goody, R.S, Itzen, A. | | Deposit date: | 2009-09-23 | | Release date: | 2010-01-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RabGDI displacement by DrrA from Legionella is a consequence of its guanine nucleotide exchange activity.
Mol.Cell, 36, 2009
|
|
3JZA
 
 | | Crystal structure of human Rab1b in complex with the GEF domain of DrrA/SidM from Legionella pneumophila | | Descriptor: | PHOSPHATE ION, Ras-related protein Rab-1B, Uncharacterized protein DrrA | | Authors: | Schoebel, S, Oesterlin, L.K, Blankenfeldt, W, Goody, R.S, Itzen, A. | | Deposit date: | 2009-09-23 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RabGDI displacement by DrrA from Legionella is a consequence of its guanine nucleotide exchange activity.
Mol.Cell, 36, 2009
|
|
3N6O
 
 | | Crystal structure of the GEF and P4M domain of DrrA/SidM from Legionella pneumophila | | Descriptor: | SULFATE ION, guanine nucleotide exchange factor | | Authors: | Schoebel, S, Blankenfeldt, W, Goody, R.S, Itzen, A. | | Deposit date: | 2010-05-26 | | Release date: | 2010-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High-affinity binding of phosphatidylinositol 4-phosphate by Legionella pneumophila DrrA.
Embo Rep., 11, 2010
|
|
3TNF
 
 | | LidA from Legionella in complex with active Rab8a | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, LidA, MAGNESIUM ION, ... | | Authors: | Schoebel, S, Cichy, A.L, Goody, R.S, Itzen, A. | | Deposit date: | 2011-09-01 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein LidA from Legionella is a Rab GTPase supereffector.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5V7V
 
 | | Cryo-EM structure of ERAD-associated E3 ubiquitin-protein ligase component HRD3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ERAD-associated E3 ubiquitin-protein ligase component HRD3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Mi, W, Schoebel, S, Stein, A, Rapoport, T.A, Liao, M. | | Deposit date: | 2017-03-20 | | Release date: | 2017-08-16 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3.
Nature, 548, 2017
|
|
6QIM
 
 | | Structure of AtPIP2;4 | | Descriptor: | Probable aquaporin PIP2-4 | | Authors: | Schoebel, S, Wang, H. | | Deposit date: | 2019-01-21 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Characterization of aquaporin-driven hydrogen peroxide transport.
Biochim Biophys Acta Biomembr, 1862, 2020
|
|
1EOE
 
 | | CRYSTAL STRUCTURE OF THE V135R MUTANT OF A SHAKER T1 DOMAIN | | Descriptor: | POTASSIUM CHANNEL KV1.1 | | Authors: | Nanao, M.H, Cushman, S.J, Jahng, A.W, DeRubeis, D, Choe, S, Pfaffinger, P.J. | | Deposit date: | 2000-03-22 | | Release date: | 2000-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Voltage dependent activation of potassium channels is coupled to T1 domain structure.
Nat.Struct.Biol., 7, 2000
|
|
1EOD
 
 | | CRYSTAL STRUCTURE OF THE N136D MUTANT OF A SHAKER T1 DOMAIN | | Descriptor: | POTASSIUM CHANNEL KV1.1 | | Authors: | Nanao, M.H, Cushman, S.J, Jahng, A.W, DeRubeis, D, Choe, S, Pfaffinger, P.J. | | Deposit date: | 2000-03-22 | | Release date: | 2000-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Voltage dependent activation of potassium channels is coupled to T1 domain structure.
Nat.Struct.Biol., 7, 2000
|
|
1NN7
 
 | |
1H1V
 
 | | gelsolin G4-G6/actin complex | | Descriptor: | ACTIN, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Choe, H, Burtnick, L.D, Mejillano, M, Yin, H.L, Robinson, R.C, Choe, S. | | Deposit date: | 2002-07-23 | | Release date: | 2003-01-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The Calcium Activation of Gelsolin:Insights from the 3A Structure of the G4-G6/Actin Complex
J.Mol.Biol., 324, 2002
|
|
5LAO
 
 | | S-nitrosylated 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Maslennikov, I, Kwiatkowski, W, Choe, S, Lipton, S.A, Guntert, P, Riek, R. | | Deposit date: | 2016-06-14 | | Release date: | 2016-08-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
5LAM
 
 | | Refined 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Maslennikov, I, Kwiatkowski, W, Choe, S, Lipton, S.A, Guntert, P, Riek, R. | | Deposit date: | 2016-06-14 | | Release date: | 2016-08-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
4DJB
 
 | | A Structural Basis for the Assembly and Functions of a Viral Polymer that Inactivates Multiple Tumor Suppressors | | Descriptor: | E4-ORF3 | | Authors: | Ou, H.D, Kwiatkowski, W, Deerinck, T.J, Noske, A, Blain, K.Y, Land, H.S, Soria, C, Powers, C.J, May, A.P, Shu, X, Tsien, R.Y, Fitzpatrick, J.A.J, Long, J.A, Ellisman, M.H, Choe, S, O'Shea, C.C. | | Deposit date: | 2012-02-01 | | Release date: | 2012-10-31 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | A Structural Basis for the Assembly and Functions of a Viral Polymer that Inactivates Multiple Tumor Suppressors.
Cell(Cambridge,Mass.), 151, 2012
|
|
5HBL
 
 | | Native rhodanese domain of YgaP prepared with 1mM DDT is S-nitrosylated | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Kwiatkowski, W, Maslennikov, I, Choe, S, Lipton, S.A, Riek, R. | | Deposit date: | 2015-12-31 | | Release date: | 2016-08-10 | | Last modified: | 2021-09-08 | | Method: | X-RAY DIFFRACTION (1.617 Å) | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
5HBP
 
 | | The crystal of rhodanese domain of YgaP treated with SNOC | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Kwiatkowski, W, Maslennikov, I, Choe, S, Lipton, S.A, Riek, R. | | Deposit date: | 2016-01-01 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
5HBQ
 
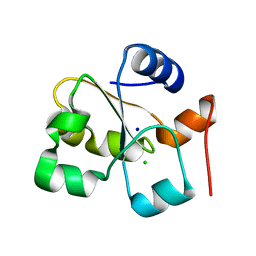 | | C63D mutant of the rhodanese domain of YgaP | | Descriptor: | CHLORIDE ION, Inner membrane protein YgaP, SODIUM ION | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Kwiatkowski, W, Maslennikov, I, Choe, S, Lipton, S.A, Riek, R. | | Deposit date: | 2016-01-02 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
5HBO
 
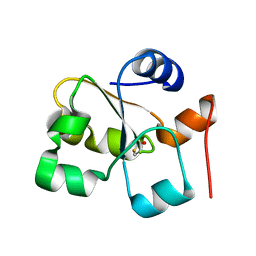 | | Native rhodanese domain of YgaP prepared without DDT is both S-nitrosylated and S-sulfhydrated | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Kwiatkowski, W, Maslennikov, I, Choe, S, Lipton, S.A, Riek, R. | | Deposit date: | 2016-01-01 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
3Q41
 
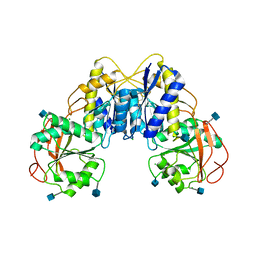 | | Crystal structure of the GluN1 N-terminal domain (NTD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Glutamate [NMDA] receptor subunit zeta-1 | | Authors: | Farina, A.N, Blain, K.Y, Maruo, T, Kwiatkowski, W, Choe, S, Nakagawa, T. | | Deposit date: | 2010-12-22 | | Release date: | 2011-03-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Separation of Domain Contacts Is Required for Heterotetrameric Assembly of Functional NMDA Receptors.
J.Neurosci., 31, 2011
|
|
3QDO
 
 | |
2WVN
 
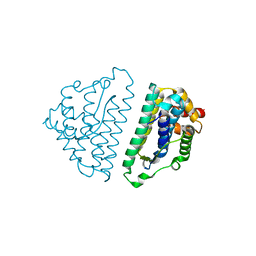 | | Structure of the HET-s N-terminal domain | | Descriptor: | SMALL S PROTEIN | | Authors: | Greenwald, J, Buhtz, C, Ritter, C, Kwiatkowski, W, Choe, S, Saupe, S.J, Riek, R. | | Deposit date: | 2009-10-19 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The Mechanism of Prion Inhibition by Het-S.
Mol.Cell, 38, 2010
|
|
2WVQ
 
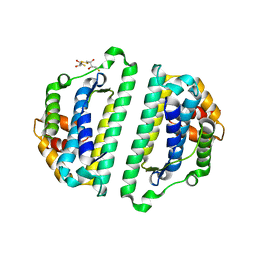 | | Structure of the HET-s N-terminal domain. Mutant D23A, P33H | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, SMALL S PROTEIN | | Authors: | Greenwald, J, Buhtz, C, Ritter, C, Kwiatkowski, W, Choe, S, Saupe, S.J, Riek, R. | | Deposit date: | 2009-10-19 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The mechanism of prion inhibition by HET-S.
Mol. Cell, 38, 2010
|
|
2WVO
 
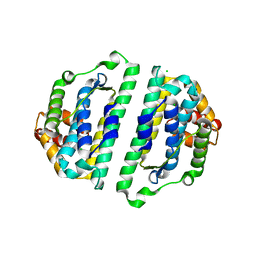 | | Structure of the HET-S N-terminal domain | | Descriptor: | CHLORIDE ION, SMALL S PROTEIN | | Authors: | Greenwald, J, Buhtz, C, Ritter, C, Kwiatkowski, W, Choe, S, Saupe, S.J, Riek, R. | | Deposit date: | 2009-10-19 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Mechanism of Prion Inhibition by Het-S.
Mol.Cell, 38, 2010
|
|
3JZ3
 
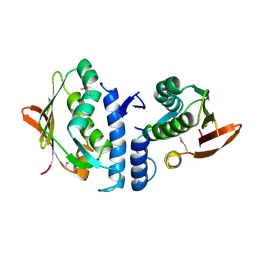 | | Structure of the cytoplasmic segment of histidine kinase QseC | | Descriptor: | SULFATE ION, Sensor protein qseC | | Authors: | Xie, W, Kwiatkowski, W, Choe, S, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2009-09-22 | | Release date: | 2010-07-21 | | Last modified: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Cytoplasmic Segment of Histidine Kinase Receptor QseC, a Key Player in Bacterial Virulence.
Protein Pept.Lett., 17, 2010
|
|
3K19
 
 | | OmpF porin | | Descriptor: | Outer membrane protein F | | Authors: | Kefala, G, Ahn, C, Krupa, M, Maslennikov, I, Kwiatkowski, W, Choe, S, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2009-09-26 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | Structures of the OmpF porin crystallized in the presence of foscholine-12.
Protein Sci., 19, 2010
|
|
